Profiling of the viable bacterial and fungal microbiota in fermented feeds using single-molecule real-time sequencing
- PMID: 32017844
- PMCID: PMC7036599
- DOI: 10.1093/jas/skaa029
Profiling of the viable bacterial and fungal microbiota in fermented feeds using single-molecule real-time sequencing
Abstract
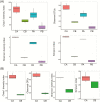
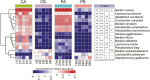
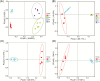
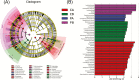
Fermented concentrated feed has been widely recognized as an ideal feed in the animal industry. In this study, we used a powerful method, coupling propidium monoazide (PMA) pretreatment with single-molecule real-time (SMRT) sequencing technology to compare the bacterial and fungal composition of feeds before and after fermentation with four added lactic acid bacteria (LAB) inoculants (one Lactobacillus casei strain and three L. plantarum strains). Five feed samples consisting of corn, soybean meal, and wheat bran were fermented with LAB additives for 3 d. Following anaerobic fermentation, the pH rapidly decreased, and the mean numbers of LAB increased from 106 to 109 colony-forming units (cfu)/g fresh matter. SMRT sequencing results showed that the abundance and diversity of bacteria and fungi in the feed were significantly higher before fermentation than after fermentation. Fifteen bacterial species and eight fungal genera were significantly altered following fermentation, and L. plantarum was the dominant species (relative abundance 88.94%) in the post-fermentation group. PMA treatment revealed that the bacteria Bacillus cereus, B. circulans, Alkaliphilus oremlandii, Cronobacter sakazakii, Paenibacillus barcinonensis, and P. amylolyticus (relative abundance >1%) were viable in the raw feed. After fermentation, their relative abundances decreased sharply to <0.2%; however, viable L. plantarum was still the dominant species post fermentation. We inferred that our LAB additives grew rapidly and inhibited harmful microorganisms and further improved feed quality. In addition, coupling PMA treatment with the Pacific Biosciences SMRT sequencing technology was a powerful tool for providing accurate live microbiota profiling data in this study.
Keywords: bacterial and fungal community; fermented feeds; lactic acid bacteria; propidium monoazide treatment; single-molecule real-time sequencing.
© The Author(s) 2020. Published by Oxford University Press on behalf of the American Society of Animal Science. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Amado I. R., Fuciños C., Fajardo P., Guerra N. P., and Pastrana L.. . 2012. Evaluation of two bacteriocin-producing probiotic lactic acid bacteria as inoculants for controlling listeria monocytogenes in grass and maize silages. Animal Feed Sci. Technol. 175(3–4):137–149. doi: 10.1016/j.anifeedsci.2012.05.006 - DOI
-
- Bao W., Mi Z., Xu H., Zheng Y., Lai Y. K., Zhang H., and Zhang W.. . 2016. Assessing quality of Medicago sativa silage by monitoring bacterial composition with single molecule, real-time sequencing technology and various physiological parameters. Sci. Reports 6:28358. doi: 10.1038/srep28358 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

