Variable impact of conformationally distinct DNA lesions on nucleosome structure and dynamics: Implications for nucleotide excision repair
- PMID: 32018112
- PMCID: PMC7060790
- DOI: 10.1016/j.dnarep.2019.102768
Variable impact of conformationally distinct DNA lesions on nucleosome structure and dynamics: Implications for nucleotide excision repair
Abstract
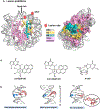
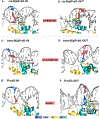
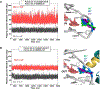
The packaging of DNA in nucleosomes presents a barrier for biological transactions including replication, transcription and repair. However, despite years of research, how the DNA is freed from the histone proteins and thereby allows the molecular machines to access the DNA remains poorly understood. We are interested in global genomic nucleotide excision repair (GG-NER). It is established that the histones are obstacles to this process, and DNA lesions are repaired less efficiently in nucleosomes than in free DNA. In the present study, we utilized molecular dynamics simulations to elucidate the nature of the distortions and dynamics imposed in the nucleosome by a set of three structually different lesions that vary in GG-NER efficiencies in free DNA, and in nucleosomes [Shafirovich, Geacintov, et. al, 2019]. Two of these are bulky lesions derived from metabolic activation of the environmental carcinogen benzo[a]pyrene, the 10R (+)-cis-anti-B[a]P-N2-dG and the stereoisomeric 10S (+)-trans-anti-B[a]P-N2-dG, which respectively adopt base-displaced/intercalated and minor groove-aligned conformations in DNA. The third is a non-bulky lesion, the 5'R-8-cyclo-2'-deoxyguanosine cross-link, produced by reactive oxygen and nitrogen species; cyclopurine lesions are highly mutagenic. These adducts are placed near the dyad axis, and rotationally with the lesion-containing strand facing towards or away from the histones. While each lesion has distinct conformational characteristics that are retained in the nucleosome, a spectrum of structural and dynamic disturbances, from slight to substantial, are displayed that depend on the lesion's structure and position in the nucleosome. We hypothesize that these intrinsic structural and dynamic distinctions provide different signals to initiate the cascade of chromatin-opening processes, including acetylation and other post translational modifications, remodeling by ATP-dependent complexes and spontaneous unwrapping that regulate the rate of access to the lesion; this may translate ultimately into varying GG-NER efficiencies, including repair resistance when signals for access are too weak.
Keywords: 5’R-8-cyclo-2’-deoxyguanosine cross-link; Benzo[a]pyrene diol epoxide-derived DNA adducts; DNA damage; Molecular dynamics; Nucleosome; Nucleotide excision repair; Rotational and translational setting.
Copyright © 2019 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no competing financial interest.
Figures
Similar articles
-
Synergistic effects of H3 and H4 nucleosome tails on structure and dynamics of a lesion-containing DNA: Binding of a displaced lesion partner base to the H3 tail for GG-NER recognition.DNA Repair (Amst). 2018 May;65:73-78. doi: 10.1016/j.dnarep.2018.02.009. Epub 2018 Mar 8. DNA Repair (Amst). 2018. PMID: 29631253 Free PMC article.
-
Nucleosome Histone Tail Conformation and Dynamics: Impacts of Lysine Acetylation and a Nearby Minor Groove Benzo[a]pyrene-Derived Lesion.Biochemistry. 2017 Apr 11;56(14):1963-1973. doi: 10.1021/acs.biochem.6b01208. Epub 2017 Mar 22. Biochemistry. 2017. PMID: 28304160 Free PMC article.
-
Nucleotide Excision Repair and Impact of Site-Specific 5',8-Cyclopurine and Bulky DNA Lesions on the Physical Properties of Nucleosomes.Biochemistry. 2019 Feb 12;58(6):561-574. doi: 10.1021/acs.biochem.8b01066. Epub 2019 Jan 7. Biochemistry. 2019. PMID: 30570250 Free PMC article.
-
Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage.DNA Repair (Amst). 2011 Feb 7;10(2):119-25. doi: 10.1016/j.dnarep.2010.10.010. Epub 2010 Dec 3. DNA Repair (Amst). 2011. PMID: 21130713 Review.
-
Nucleotide excision repair leaves a mark on chromatin: DNA damage detection in nucleosomes.Cell Mol Life Sci. 2021 Dec;78(24):7925-7942. doi: 10.1007/s00018-021-03984-7. Epub 2021 Nov 3. Cell Mol Life Sci. 2021. PMID: 34731255 Free PMC article. Review.
Cited by
-
Targeting the DNA Damage Response for Cancer Therapy.Int J Mol Sci. 2023 Nov 2;24(21):15907. doi: 10.3390/ijms242115907. Int J Mol Sci. 2023. PMID: 37958890 Free PMC article. Review.
-
Recent Advances in Investigating Functional Dynamics of Chromatin.Front Genet. 2022 Apr 5;13:870640. doi: 10.3389/fgene.2022.870640. eCollection 2022. Front Genet. 2022. PMID: 35450211 Free PMC article. Review.
-
DNA damage and repair in the nucleosome: insights from computational methods.Biophys Rev. 2024 Mar 12;16(3):345-356. doi: 10.1007/s12551-024-01183-9. eCollection 2024 Jun. Biophys Rev. 2024. PMID: 39099841 Free PMC article. Review.
-
Molecular dynamics simulations reveal how H3K56 acetylation impacts nucleosome structure to promote DNA exposure for lesion sensing.DNA Repair (Amst). 2021 Nov;107:103201. doi: 10.1016/j.dnarep.2021.103201. Epub 2021 Aug 8. DNA Repair (Amst). 2021. PMID: 34399316 Free PMC article.
-
DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy.Signal Transduct Target Ther. 2021 Jul 9;6(1):254. doi: 10.1038/s41392-021-00648-7. Signal Transduct Target Ther. 2021. PMID: 34238917 Free PMC article. Review.
References
-
- Wang ZG, Wu XH, Friedberg EC, Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes, J. Biol. Chem, 266 (1991) 22472–22478. - PubMed
-
- Smerdon MJ, Conconi A, Modulation of DNA damage and DNA repair in chromatin, Prog. Nucleic Acid Res. Mol. Biol, 62 (1999) 227–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

