A Unique Regulation Region in the 3' UTR of HLA-G with a Promising Potential
- PMID: 32019184
- PMCID: PMC7037441
- DOI: 10.3390/ijms21030900
A Unique Regulation Region in the 3' UTR of HLA-G with a Promising Potential
Abstract
Human leukocyte antigen G (HLA-G) is a non-classical human leukocyte antigen (HLA) class I protein that interacts with inhibitory receptors and is commonly overexpressed in various cancers, thereby establishing itself as an inhibitory checkpoint immune ligand. It is also expressed in trophoblast cells during pregnancy and protects the fetus from immune rejection. Despite its crucial role and its intriguing expression pattern, the regulation of HLA-G's expression is only partially understood. HLA-G's mRNA is expressed in many tissues but the protein expression is restricted only to the cells mentioned above. Therefore, we suggest that HLA-G is post-transcriptionally regulated. Here, we reveal a distinctive site present only in the 3' Untranslated region (UTR) of HLA-G, which might explain its unique expression pattern. Consequently, we attempted to find binding factors such as RNA binding proteins (RBPs) and microRNAS (miRs) that regulate HLA-G expression by interacting with this distinct site present in its 3' UTR. Our research indicates that this site should be further studied in order to reveal its significance.
Keywords: 3′ UTR; HLA-G; RNA binding proteins; microRNA.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

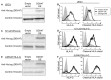
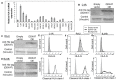

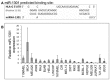
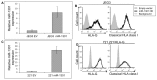

References
-
- Carosella E.D., Rouas-Freiss N., Roux D.T., LeMoreau P., Le Maoult J. Advances in Immunology. Volume 127. Academic Press Inc.; Cambridge, MA, USA: 2015. HLA-G. An Immune Checkpoint Molecule; pp. 33–144. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

