Let-7 microRNA is a critical regulator in controlling the growth and function of silk gland in the silkworm
- PMID: 32019402
- PMCID: PMC7237193
- DOI: 10.1080/15476286.2020.1726128
Let-7 microRNA is a critical regulator in controlling the growth and function of silk gland in the silkworm
Abstract
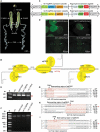
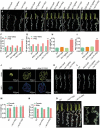
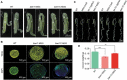
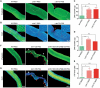
The silk gland is characterized by high protein synthesis. However, the molecular mechanisms controlling silk gland growth and silk protein synthesis remain undetermined. Here we demonstrated that CRISPR/Cas9-based knockdown of let-7 or the whole cluster promoted endoreduplication and enlargement of the silk gland, accompanied by changing silk yield, whereas transgenic overexpression of let-7 led to atrophy and degeneration of the silk gland. Mechanistically, let-7 controls cell growth in the silk gland through coordinating nutrient metabolism processes and energy signalling pathways. Transgenic overexpression of pyruvate carboxylase, a novel target of let-7, resulted in enlargement of the silk glands, which is consistent with the abnormal phenotype of the let-7 knockdown. Overall, our data reveal a previously unknown miRNA-mediated regulation of silk gland growth and physiology and shed light on involvement of let-7 as a critical stabilizer and booster in carbohydrate metabolism, which may have important implications for understanding of the molecular mechanism and physiological function of specialized organs in other species.
Keywords: CRISP/Cas9; Let-7 microRNA; Silk gland; endoreduplication; glycometabolism; pyruvate carboxylase.
Figures


Similar articles
-
MicroRNA let-7 targets BmCDK1 to regulate cell proliferation and endomitosis of silk gland in the silkworm, Bombyx mori.Insect Sci. 2024 Aug;31(4):1026-1040. doi: 10.1111/1744-7917.13302. Epub 2023 Dec 5. Insect Sci. 2024. PMID: 38053466
-
Let-7 microRNA targets BmCentrin to modulate the development and functionality of the middle silk gland in the silkworm, Bombyx mori.Insect Sci. 2025 Feb;32(1):95-114. doi: 10.1111/1744-7917.13380. Epub 2024 May 29. Insect Sci. 2025. PMID: 38812265
-
CRISPR/Cas9-mediated gene editing of the let-7 seed sequence improves silk yield in the silkworm, Bombyx mori.Int J Biol Macromol. 2023 Jul 15;243:124793. doi: 10.1016/j.ijbiomac.2023.124793. Epub 2023 May 12. Int J Biol Macromol. 2023. PMID: 37182624
-
The advances and perspectives of recombinant protein production in the silk gland of silkworm Bombyx mori.Transgenic Res. 2014 Oct;23(5):697-706. doi: 10.1007/s11248-014-9826-8. Epub 2014 Aug 12. Transgenic Res. 2014. PMID: 25113390 Review.
-
Transgenic silkworms that weave recombinant proteins into silk cocoons.Biotechnol Lett. 2011 Apr;33(4):645-54. doi: 10.1007/s10529-010-0498-z. Epub 2010 Dec 24. Biotechnol Lett. 2011. PMID: 21184136 Review.
Cited by
-
Long Noncoding RNA lncR17454 Regulates Metamorphosis of Silkworm Through let-7 miRNA Cluster.J Insect Sci. 2022 May 1;22(3):12. doi: 10.1093/jisesa/ieac028. J Insect Sci. 2022. PMID: 35640247 Free PMC article.
-
20-Hydroxyecdysone-responsive microRNAs of insects.RNA Biol. 2020 Oct;17(10):1454-1471. doi: 10.1080/15476286.2020.1775395. Epub 2020 Jun 16. RNA Biol. 2020. PMID: 32482109 Free PMC article.
-
Age and Behavior-Dependent Differential miRNAs Expression in the Hypopharyngeal Glands of Honeybees (Apis mellifera L.).Insects. 2021 Aug 26;12(9):764. doi: 10.3390/insects12090764. Insects. 2021. PMID: 34564204 Free PMC article.
-
Decoding the regulatory roles of non-coding RNAs in cellular metabolism and disease.Mol Ther. 2023 Jun 7;31(6):1562-1576. doi: 10.1016/j.ymthe.2023.04.012. Epub 2023 Apr 27. Mol Ther. 2023. PMID: 37113055 Free PMC article. Review.
-
Combined analysis of the proteome and metabolome provides insight into microRNA-1174 function in Aedes aegypti mosquitoes.Parasit Vectors. 2023 Aug 9;16(1):271. doi: 10.1186/s13071-023-05859-1. Parasit Vectors. 2023. PMID: 37559132 Free PMC article.
References
-
- Akai H. The structure and ultrastructure of the silk gland. Experientia. 1983;39:443–449.
-
- Xia Q, Li S, Feng Q. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu Rev Entomol. 2014;59:513–536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
