The Toxoplasma gondii Cyst Wall Interactome
- PMID: 32019789
- PMCID: PMC7002340
- DOI: 10.1128/mBio.02699-19
The Toxoplasma gondii Cyst Wall Interactome
Abstract
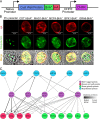
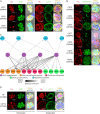
A characteristic of the latent cyst stage of Toxoplasma gondii is a thick cyst wall that forms underneath the membrane of the bradyzoite vacuole. Previously, our laboratory group published a proteomic analysis of purified in vitro cyst wall fragments that identified an inventory of cyst wall components. To further refine our understanding of the composition of the cyst wall, several cyst wall proteins were tagged with a promiscuous biotin ligase (BirA*), and their interacting partners were screened by streptavidin affinity purification. Within the cyst wall pulldowns, previously described cyst wall proteins, dense granule proteins, and uncharacterized hypothetical proteins were identified. Several of the newly identified hypothetical proteins were validated to be novel components of the cyst wall and tagged with BirA* to expand the model of the cyst wall interactome. Community detection of the cyst wall interactome model revealed three distinct clusters: a dense granule, a cyst matrix, and a cyst wall cluster. Characterization of several of the identified cyst wall proteins using genetic strategies revealed that MCP3 affects in vivo cyst sizes. This study provides a model of the potential protein interactions within the cyst wall and the groundwork to understand cyst wall formation.IMPORTANCE A model of the cyst wall interactome was constructed using proteins identified through BioID. The proteins within this cyst wall interactome model encompass several proteins identified in a prior characterization of the cyst wall proteome. This model provides a more comprehensive understanding of the composition of the cyst wall and may lead to insights on how the cyst wall is formed.
Keywords: BirA; Toxoplasma gondii; cyst wall; protein interactions; proteomics.
Copyright © 2020 Tu et al.
Figures



References
-
- Buchholz KR, Fritz HM, Chen X, Durbin-Johnson B, Rocke DM, Ferguson DJ, Conrad PA, Boothroyd JC. 2011. Identification of tissue cyst wall components by transcriptome analysis of in vivo and in vitro Toxoplasma gondii bradyzoites. Eukaryot Cell 10:1637–1647. doi:10.1128/EC.05182-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

