A CLOCK-binding small molecule disrupts the interaction between CLOCK and BMAL1 and enhances circadian rhythm amplitude
- PMID: 32019867
- PMCID: PMC7076206
- DOI: 10.1074/jbc.RA119.011332
A CLOCK-binding small molecule disrupts the interaction between CLOCK and BMAL1 and enhances circadian rhythm amplitude
Abstract
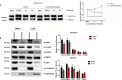
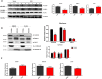
Proper function of many physiological processes requires a robust circadian clock. Disruptions of the circadian clock can result in metabolic diseases, mood disorders, and accelerated aging. Therefore, identifying small molecules that specifically modulate regulatory core clock proteins may potentially enable better management of these disorders. In this study, we applied a structure-based molecular-docking approach to find small molecules that specifically bind to the core circadian regulator, the transcription factor circadian locomotor output cycles kaput (CLOCK). We identified 100 candidate molecules by virtual screening of ∼2 million small molecules for those predicted to bind closely to the interface in CLOCK that interacts with its transcriptional co-regulator, Brain and muscle Arnt-like protein-1 (BMAL1). Using a mammalian two-hybrid system, real-time monitoring of circadian rhythm in U2OS cells, and various biochemical assays, we tested these compounds experimentally and found one, named CLK8, that specifically bound to and interfered with CLOCK activity. We show that CLK8 disrupts the interaction between CLOCK and BMAL1 and interferes with nuclear translocation of CLOCK both in vivo and in vitro Results from further experiments indicated that CLK8 enhances the amplitude of the cellular circadian rhythm by stabilizing the negative arm of the transcription/translation feedback loop without affecting period length. Our results reveal CLK8 as a tool for further studies of CLOCK's role in circadian rhythm amplitude regulation and as a potential candidate for therapeutic development to manage disorders associated with dampened circadian rhythms.
Keywords: brain and muscle Arnt-like protein-1 (BMAL1); circadian clock; circadian locomotor output cycles kaput (CLOCK); circadian regulation; circadian rhythm amplitude; drug design; drug development; gene expression; gene regulation; protein expression; transcription; transcription coregulator.
© 2020 Doruk et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

