TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages
- PMID: 32019932
- PMCID: PMC7000671
- DOI: 10.1038/s41467-020-14508-5
TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages
Abstract
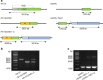
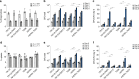
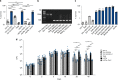
Mycobacterium tuberculosis (Mtb) strains are classified into different phylogenetic lineages (L), three of which (L2/L3/L4) emerged from a common progenitor after the loss of the MmpS6/MmpL6-encoding Mtb-specific deletion 1 region (TbD1). These TbD1-deleted "modern" lineages are responsible for globally-spread tuberculosis epidemics, whereas TbD1-intact "ancestral" lineages tend to be restricted to specific geographical areas, such as South India and South East Asia (L1) or East Africa (L7). By constructing and characterizing a panel of recombinant TbD1-knock-in and knock-out strains and comparison with clinical isolates, here we show that deletion of TbD1 confers to Mtb a significant increase in resistance to oxidative stress and hypoxia, which correlates with enhanced virulence in selected cellular, guinea pig and C3HeB/FeJ mouse infection models, the latter two mirroring in part the development of hypoxic granulomas in human disease progression. Our results suggest that loss of TbD1 at the origin of the L2/L3/L4 Mtb lineages was a key driver for their global epidemic spread and outstanding evolutionary success.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The TbD1 Locus Mediates a Hypoxia-Induced Copper Response in Mycobacterium bovis.Front Microbiol. 2022 Apr 14;13:817952. doi: 10.3389/fmicb.2022.817952. eCollection 2022. Front Microbiol. 2022. PMID: 35495699 Free PMC article.
-
A new evolutionary scenario for the Mycobacterium tuberculosis complex.Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3684-9. doi: 10.1073/pnas.052548299. Epub 2002 Mar 12. Proc Natl Acad Sci U S A. 2002. PMID: 11891304 Free PMC article.
-
High proportion of modern genotypes of M. tuberculosis and their affinity with drug resistance in northern region of India.J Glob Antimicrob Resist. 2017 Sep;10:84-87. doi: 10.1016/j.jgar.2017.04.010. Epub 2017 Jul 17. J Glob Antimicrob Resist. 2017. PMID: 28729202
-
Molecular epidemiology of M. tuberculosis in Ethiopia: A systematic review and meta-analysis.Tuberculosis (Edinb). 2019 Sep;118:101858. doi: 10.1016/j.tube.2019.101858. Epub 2019 Aug 7. Tuberculosis (Edinb). 2019. PMID: 31430694 Free PMC article.
-
Host-pathogen coevolution in human tuberculosis.Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):850-9. doi: 10.1098/rstb.2011.0316. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22312052 Free PMC article. Review.
Cited by
-
Lineage-Specific Proteomic Signatures in the Mycobacterium tuberculosis Complex Reveal Differential Abundance of Proteins Involved in Virulence, DNA Repair, CRISPR-Cas, Bioenergetics and Lipid Metabolism.Front Microbiol. 2020 Sep 22;11:550760. doi: 10.3389/fmicb.2020.550760. eCollection 2020. Front Microbiol. 2020. PMID: 33072011 Free PMC article.
-
An imputed ancestral reference genome for the Mycobacterium tuberculosis complex better captures structural genomic diversity for reference-based alignment workflows.Microb Genom. 2024 Jan;10(1):001165. doi: 10.1099/mgen.0.001165. Microb Genom. 2024. PMID: 38175684 Free PMC article.
-
Underwhelming or Misunderstood? Genetic Variability of Pattern Recognition Receptors in Immune Responses and Resistance to Mycobacterium tuberculosis.Front Immunol. 2021 Jun 30;12:714808. doi: 10.3389/fimmu.2021.714808. eCollection 2021. Front Immunol. 2021. PMID: 34276708 Free PMC article. Review.
-
The TbD1 Locus Mediates a Hypoxia-Induced Copper Response in Mycobacterium bovis.Front Microbiol. 2022 Apr 14;13:817952. doi: 10.3389/fmicb.2022.817952. eCollection 2022. Front Microbiol. 2022. PMID: 35495699 Free PMC article.
-
Whole genome sequencing-based identification of human tuberculosis caused by animal-lineage Mycobacterium orygis.J Clin Microbiol. 2023 Nov 21;61(11):e0026023. doi: 10.1128/jcm.00260-23. Epub 2023 Oct 25. J Clin Microbiol. 2023. PMID: 37877705 Free PMC article.
References
-
- WHO. Global tuberculosis report 2018. Geneva: World Health Organisation; 2019.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

