Transcriptome Analysis Reveals the Resistance Mechanism of Pseudomonas aeruginosa to Tachyplesin I
- PMID: 32021330
- PMCID: PMC6970625
- DOI: 10.2147/IDR.S226687
Transcriptome Analysis Reveals the Resistance Mechanism of Pseudomonas aeruginosa to Tachyplesin I
Abstract
Background: Tachyplesin I is a cationic antimicrobial peptide with a typical cyclic antiparallel β-sheet structure. We previously demonstrated that long-term continuous exposure to increased concentration of tachyplesin I can induce resistant Gram-negative bacteria. However, no significant information is available about the resistance mechanism of Pseudomonas aeruginosa (P. aeruginosa) to tachyplesin I.
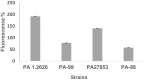
Materials and methods: In this study, the global gene expression profiling of P. aeruginosa strain PA-99 and P. aeruginosa CGMCC1.2620 (PA1.2620) was conducted using transcriptome sequencing. For this purpose, outer membrane permeability and outer membrane proteins (OMPs) were further analyzed.
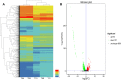
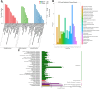
Results: Transcriptome sequencing detected 672 upregulated and 787 downregulated genes, covering Clusters of Orthologous Groups (COGs) of P. aeruginosa strain PA-99 compared with PA1.2620. Totally, 749 differentially expressed genes (DEGs) were assigned to 98 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and among them, a two-component regulatory system, a beta-lactam resistance system, etc. were involved in some known genes resistant to drugs. Additionally, we further attempted to indicate whether the resistance mechanism of P. aeruginosa to tachyplesin I was associated with the changes of outer membrane permeability and OMPs.
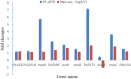
Conclusion: Our results indicated that P. aeruginosa resistant to tachyplesin I was mainly related to reduced entry of tachyplesin I into the bacterial cell due to overexpression of efflux pump, in addition to a decrease of outer membrane permeability. Our findings were also validated by pathway enrichment analysis and quantitative reverse transcription polymerase chain reaction (RT-qPCR). This study may provide a promising guidance for understanding the resistance mechanism of P. aeruginosa to tachyplesin I.
Keywords: P. aeruginosa; antibiotic resistance; differentially expressed genes; outer membrane permeability; outer membrane proteins; tachyplesin I.
© 2020 Hong et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Co-expression Mechanism Analysis of Different Tachyplesin I-Resistant Strains in Pseudomonas aeruginosa Based on Transcriptome Sequencing.Front Microbiol. 2022 Apr 7;13:871290. doi: 10.3389/fmicb.2022.871290. eCollection 2022. Front Microbiol. 2022. PMID: 35464984 Free PMC article.
-
Experimental Induction of Bacterial Resistance to the Antimicrobial Peptide Tachyplesin I and Investigation of the Resistance Mechanisms.Antimicrob Agents Chemother. 2016 Sep 23;60(10):6067-75. doi: 10.1128/AAC.00640-16. Print 2016 Oct. Antimicrob Agents Chemother. 2016. PMID: 27480861 Free PMC article.
-
Tachyplesin Causes Membrane Instability That Kills Multidrug-Resistant Bacteria by Inhibiting the 3-Ketoacyl Carrier Protein Reductase FabG.Front Microbiol. 2018 May 1;9:825. doi: 10.3389/fmicb.2018.00825. eCollection 2018. Front Microbiol. 2018. PMID: 29765362 Free PMC article.
-
Comparative transcription analysis of resistant mutants against four different antibiotics in Pseudomonas aeruginosa.Microb Pathog. 2021 Nov;160:105166. doi: 10.1016/j.micpath.2021.105166. Epub 2021 Sep 1. Microb Pathog. 2021. PMID: 34480983
-
Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa.Rev Infect Dis. 1988 Jul-Aug;10(4):770-5. doi: 10.1093/clinids/10.4.770. Rev Infect Dis. 1988. PMID: 2460909 Review.
Cited by
-
Heterogeneous efflux pump expression underpins phenotypic resistance to antimicrobial peptides.Elife. 2025 Jul 3;13:RP99752. doi: 10.7554/eLife.99752. Elife. 2025. PMID: 40607907 Free PMC article.
-
Co-expression Mechanism Analysis of Different Tachyplesin I-Resistant Strains in Pseudomonas aeruginosa Based on Transcriptome Sequencing.Front Microbiol. 2022 Apr 7;13:871290. doi: 10.3389/fmicb.2022.871290. eCollection 2022. Front Microbiol. 2022. PMID: 35464984 Free PMC article.
-
Molecular basis of a bacterial-amphibian symbiosis revealed by comparative genomics, modeling, and functional testing.ISME J. 2022 Mar;16(3):788-800. doi: 10.1038/s41396-021-01121-7. Epub 2021 Oct 2. ISME J. 2022. PMID: 34601502 Free PMC article.
References
-
- Overhag J, Strehmel J, Strempel N. Potential application of antimicrobial peptides in the treatment of bacterial biofilm infections. Curr Pharm Des. 2015;21(1):66–84. - PubMed
LinkOut - more resources
Full Text Sources

