Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV
- PMID: 32022276
- PMCID: PMC7166505
- DOI: 10.1002/jmv.25698
Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV
Abstract
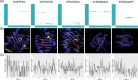
The 2019 novel coronavirus (2019-nCoV) outbreak has caused a large number of deaths with thousands of confirmed cases worldwide, especially in East Asia. This study took an immunoinformatics approach to identify significant cytotoxic T lymphocyte (CTL) and B cell epitopes in the 2019-nCoV surface glycoprotein. Also, interactions between identified CTL epitopes and their corresponding major histocompatibility complex (MHC) class I supertype representatives prevalent in China were studied by molecular dynamics simulations. We identified five CTL epitopes, three sequential B cell epitopes and five discontinuous B cell epitopes in the viral surface glycoprotein. Also, during simulations, the CTL epitopes were observed to be binding MHC class I peptide-binding grooves via multiple contacts, with continuous hydrogen bonds and salt bridge anchors, indicating their potential in generating immune responses. Some of these identified epitopes can be potential candidates for the development of 2019-nCoV vaccines.
Keywords: 2019-nCoV; COVID-19; SARS-CoV-2; coronavirus; epitope prediction; immunoinformatics.
© 2020 Wiley Periodicals, Inc.
Figures
Comment in
-
Defining protective epitopes for COVID-19 vaccination models.J Med Virol. 2020 Oct;92(10):1772-1773. doi: 10.1002/jmv.25876. Epub 2020 Jun 2. J Med Virol. 2020. PMID: 32285942 Free PMC article. No abstract available.
References
-
- World Health Organization . Novel coronavirus (2019‐nCoV) situation report‐7. Report 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2...
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

