A window into lysogeny: revealing temperate phage biology with transcriptomics
- PMID: 32022660
- PMCID: PMC7067206
- DOI: 10.1099/mgen.0.000330
A window into lysogeny: revealing temperate phage biology with transcriptomics
Abstract
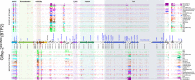
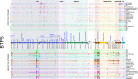
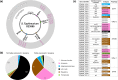
Prophages are integrated phage elements that are a pervasive feature of bacterial genomes. The fitness of bacteria is enhanced by prophages that confer beneficial functions such as virulence, stress tolerance or phage resistance, and these functions are encoded by 'accessory' or 'moron' loci. Whilst the majority of phage-encoded genes are repressed during lysogeny, accessory loci are often highly expressed. However, it is challenging to identify novel prophage accessory loci from DNA sequence data alone. Here, we use bacterial RNA-seq data to examine the transcriptional landscapes of five Salmonella prophages. We show that transcriptomic data can be used to heuristically enrich for prophage features that are highly expressed within bacterial cells and represent functionally important accessory loci. Using this approach, we identify a novel antisense RNA species in prophage BTP1, STnc6030, which mediates superinfection exclusion of phage BTP1. Bacterial transcriptomic datasets are a powerful tool to explore the molecular biology of temperate phages.
Keywords: transcriptomics; RNA-seq; bacteriophage; lysogeny; prophage.
Conflict of interest statement
Rocío Canals was employed by the University of Liverpool at the time of the study and is now an employee of the GSK group of companies.
Figures






References
-
- Ptashne M. A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

