Development of a long non-coding RNA signature for prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal adenocarcinoma
- PMID: 32023246
- PMCID: PMC7001901
- DOI: 10.1371/journal.pone.0226595
Development of a long non-coding RNA signature for prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal adenocarcinoma
Abstract
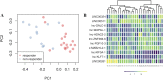
Standard treatment for locally advanced rectal adenocarcinoma (LARC) includes a combination of chemotherapy with pyrimidine analogues, such as capecitabine, and radiation therapy, followed by surgery. Currently no clinically useful genomic predictors of benefit from neoadjuvant chemoradiotherapy (nCRT) exist for LARC. In this study we assessed the expression of 8,127 long noncoding RNAs (lncRNAs), poorly studied in LARC, to infer their ability in classifying patients' pathological complete response (pCR). We collected and analyzed, using lncRNA-specific Agilent microarrays a consecutive series of 61 LARC cases undergoing nCRT. Potential lncRNA predictors in responders and non-responders to nCRT were identified with LASSO regression, and a model was optimized using k-fold cross-validation after selection of the three most informative lncRNA. 11 lncRNAs were differentially expressed with false discovery rate < 0.01 between responders and non-responders to NACT. We identified lnc-KLF7-1, lnc-MAB21L2-1, and LINC00324 as the most promising variable subset for classification building. Overall sensitivity and specificity were 0.91 and 0.94 respectively, with an AUC of our ROC curve = 0.93. Our study shows for the first time that lncRNAs can accurately predict response in LARC undergoing nCRT. Our three-lncRNA based signature must be independently validated and further analyses must be conducted to fully understand the biological role of the identified signature, but our results suggest lncRNAs may be an ideal biomarker for response prediction in the studied setting.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients.BMC Med Genomics. 2014 Dec 11;7:68. doi: 10.1186/s12920-014-0068-7. BMC Med Genomics. 2014. PMID: 25496125 Free PMC article.
-
Tumor microRNAs Identified by Small RNA Sequencing as Potential Response Predictors in Locally Advanced Rectal Cancer Patients Treated With Neoadjuvant Chemoradiotherapy.Cancer Genomics Proteomics. 2020 May-Jun;17(3):249-257. doi: 10.21873/cgp.20185. Cancer Genomics Proteomics. 2020. PMID: 32345666 Free PMC article.
-
Prediction and validation of pathologic complete response for locally advanced rectal cancer under neoadjuvant chemoradiotherapy based on a novel predictor using interpretable machine learning.Eur J Surg Oncol. 2024 Dec;50(12):108738. doi: 10.1016/j.ejso.2024.108738. Epub 2024 Oct 6. Eur J Surg Oncol. 2024. PMID: 39395242
-
Mucinous histology is a negative predictor of neoadjuvant chemoradiotherapy for locally advanced rectal adenocarcinoma.BMC Gastroenterol. 2024 Aug 13;24(1):263. doi: 10.1186/s12876-024-03359-9. BMC Gastroenterol. 2024. PMID: 39138423 Free PMC article.
-
Prognostic Value of ctDNA Detection in Patients With Locally Advanced Rectal Cancer Undergoing Neoadjuvant Chemoradiotherapy: A Systematic Review and Meta-analysis.Oncologist. 2023 Dec 11;28(12):e1198-e1208. doi: 10.1093/oncolo/oyad151. Oncologist. 2023. PMID: 37294663 Free PMC article.
Cited by
-
Construction and characterization of rectal cancer-related lncRNA-mRNA ceRNA network reveals prognostic biomarkers in rectal cancer.IET Syst Biol. 2021 Aug;15(6):192-204. doi: 10.1049/syb2.12035. Epub 2021 Oct 6. IET Syst Biol. 2021. PMID: 34613665 Free PMC article.
-
The Clinical Utility of lncRNAs and Their Application as Molecular Biomarkers in Breast Cancer.Int J Mol Sci. 2023 Apr 18;24(8):7426. doi: 10.3390/ijms24087426. Int J Mol Sci. 2023. PMID: 37108589 Free PMC article. Review.
-
The Expression Profiles of lncRNAs Are Associated with Neoadjuvant Chemotherapy Resistance in Locally Advanced, Luminal B-Type Breast Cancer.Int J Mol Sci. 2024 Jul 24;25(15):8077. doi: 10.3390/ijms25158077. Int J Mol Sci. 2024. PMID: 39125649 Free PMC article.
-
ceRNA Networks: The Backbone Role in Neoadjuvant Chemoradiotherapy Resistance/Sensitivity of Locally Advanced Rectal Cancer.Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211062313. doi: 10.1177/15330338211062313. Technol Cancer Res Treat. 2021. PMID: 34908512 Free PMC article.
-
Targeting non-coding RNAs to overcome cancer therapy resistance.Signal Transduct Target Ther. 2022 Apr 13;7(1):121. doi: 10.1038/s41392-022-00975-3. Signal Transduct Target Ther. 2022. PMID: 35418578 Free PMC article. Review.
References
-
- Sauer R, Liersch T, Merkel S, et al. Preoperative Versus Postoperative Chemoradiotherapy for Locally Advanced Rectal Cancer: Results of the German CAO/ARO/AIO-94 Randomized Phase III Trial After a Median Follow-Up of 11 Years. J Clin Oncol. 2012;30(16):1926–1933. 10.1200/JCO.2011.40.1836 - DOI - PubMed
-
- Pucciarelli S, Del Bianco P, Efficace F, et al. Patient-Reported Outcomes After Neoadjuvant Chemoradiotherapy for Rectal Cancer: A Multicenter Prospective Observational Study. Ann Surg. 2011;253(1). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

