Reporting, Epidemic Growth, and Reproduction Numbers for the 2019 Novel Coronavirus (2019-nCoV) Epidemic
- PMID: 32023340
- PMCID: PMC7077751
- DOI: 10.7326/M20-0358
Reporting, Epidemic Growth, and Reproduction Numbers for the 2019 Novel Coronavirus (2019-nCoV) Epidemic
Figures

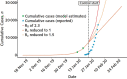
References
-
- BNO News. Tracking coronavirus: map, data and timeline. 2 February 2020. Accessed at https://bnonews.com/index.php/2020/02/the-latest-coronavirus-cases. on 3 February 2020.
-
- China Centers for Disease Control. Distribution of pneumonia in a new coronavirus infection. 2020. Accessed at http://2019ncov.chinacdc.cn/2019-nCoV. on 3 February 2020.
-
- Imai N, Cori A, Dorigatti I, et al. Report 3: transmissibility of 2019-nCoV. 25 January 2020. Accessed at www.imperial.ac.uk/mrc-global-infectious-disease-analysis/news--wuhan-co.... on 25 January 2020.
MeSH terms
LinkOut - more resources
Full Text Sources
