Oct4-Mediated Inhibition of Lsd1 Activity Promotes the Active and Primed State of Pluripotency Enhancers
- PMID: 32023463
- PMCID: PMC7316367
- DOI: 10.1016/j.celrep.2019.11.040
Oct4-Mediated Inhibition of Lsd1 Activity Promotes the Active and Primed State of Pluripotency Enhancers
Abstract
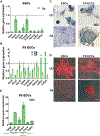
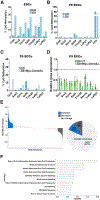
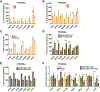
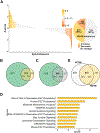
An aberrant increase in pluripotency gene (PpG) expression due to enhancer reactivation could induce stemness and enhance the tumorigenicity of cancer stem cells. Silencing of PpG enhancers (PpGe) during embryonic stem cell differentiation involves Lsd1-mediated H3K4me1 demethylation and DNA methylation. Here, we observed retention of H3K4me1 and DNA hypomethylation at PpGe associated with a partial repression of PpGs in F9 embryonal carcinoma cells (ECCs) post-differentiation. H3K4me1 demethylation in F9 ECCs could not be rescued by Lsd1 overexpression. Given our observation that H3K4me1 demethylation is accompanied by strong Oct4 repression in P19 ECCs, we tested if Oct4 interaction with Lsd1 affects its catalytic activity. Our data show a dose-dependent inhibition of Lsd1 activity by Oct4 and retention of H3K4me1 at PpGe in Oct4-overexpressing P19 ECCs. These data suggest that Lsd1-Oct4 interaction in cancer stem cells could establish a "primed" enhancer state that is susceptible to reactivation, leading to aberrant PpG expression.
Keywords: DNA methylation; Dnmt3a; Lsd1; Oct4; cancer stem cells; embryonal carcinoma cells; enhancer priming; enhancers; histone demethylation; pluripotency.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


References
-
- Alonso A, Breuer B, Steuer B, and Fischer J. (1991). The F9-EC cell line as a model for the analysis of differentiation. Int. J. Dev. Biol. 35, 389–397. - PubMed
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Andrews PW, Matin MM, Bahrami AR, Damjanov I, Gokhale P, and Draper JS (2005). Embryonic stem (ES) cells and embryonal carcinoma (EC) cells: opposite sides of the same coin. Biochem. Soc. Trans. 33, 1526–1530. - PubMed
-
- Aran D, and Hellman A. (2013). DNA methylation of transcriptional enhancers and cancer predisposition. Cell 754, 11–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

