Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression
- PMID: 32023484
- PMCID: PMC7397554
- DOI: 10.1016/j.molcel.2020.01.014
Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression
Abstract
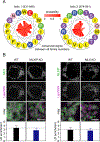
Lipid droplets (LDs) store lipids for energy and are central to cellular lipid homeostasis. The mechanisms coordinating lipid storage in LDs with cellular metabolism are unclear but relevant to obesity-related diseases. Here we utilized genome-wide screening to identify genes that modulate lipid storage in macrophages, a cell type involved in metabolic diseases. Among ∼550 identified screen hits is MLX, a basic helix-loop-helix leucine-zipper transcription factor that regulates metabolic processes. We show that MLX and glucose-sensing family members MLXIP/MondoA and MLXIPL/ChREBP bind LDs via C-terminal amphipathic helices. When LDs accumulate in cells, these transcription factors bind to LDs, reducing their availability for transcriptional activity and attenuating the response to glucose. Conversely, the absence of LDs results in hyperactivation of MLX target genes. Our findings uncover a paradigm for a lipid storage response in which binding of MLX transcription factors to LD surfaces adjusts the expression of metabolic genes to lipid storage levels.
Keywords: ChREBP; MLX; MLXIP; MLXIPL; MondoA; MondoB; glucose; lipid droplets; metabolism; transcription factor.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



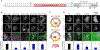

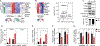
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

