Genome-Wide Identification, Expression Profile and Evolution Analysis of Karyopherin β Gene Family in Solanum tuberosum Group Phureja DM1-3 Reveals Its Roles in Abiotic Stresses
- PMID: 32023817
- PMCID: PMC7037939
- DOI: 10.3390/ijms21030931
Genome-Wide Identification, Expression Profile and Evolution Analysis of Karyopherin β Gene Family in Solanum tuberosum Group Phureja DM1-3 Reveals Its Roles in Abiotic Stresses
Abstract
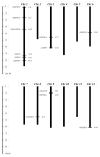
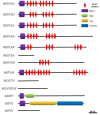
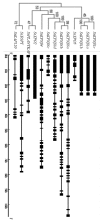
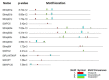
In eukaryotic cells, nucleocytoplasmic trafficking of macromolecules is largely mediated by Karyopherin β/Importin (KPNβ or Impβ) nuclear transport factors, and they import and export cargo proteins or RNAs via the nuclear pores across the nuclear envelope, consequently effecting the cellular signal cascades in response to pathogen attack and environmental cues. Although achievements on understanding the roles of several KPNβs have been obtained from model plant Arabidopsis thaliana, comprehensive analysis of potato KPNβ gene family is yet to be elucidated. In our genome-wide identifications, a total of 13 StKPNβ (Solanum tuberosum KPNβ) genes were found in the genome of the doubled monoploid S. tuberosum Group Phureja DM1-3. Sequence alignment and conserved domain analysis suggested the presence of importin-β N-terminal domain (IBN_N, PF08310) or Exporin1-like domain (XpoI, PF08389) at N-terminus and HEAT motif at the C-terminal portion in most StKPNβs. Phylogenetic analysis indicated that members of StKPNβ could be classified into 16 subgroups in accordance with their homology to human KPNβs, which was also supported by exon-intron structure, consensus motifs, and domain compositions. RNA-Seq analysis and quantitative real-time PCR experiments revealed that, except StKPNβ3d and StKPNβ4, almost all StKPNβs were ubiquitously expressed in all tissues analyzed, whereas transcriptional levels of several StKPNβs were increased upon biotic/abiotic stress or phytohormone treatments, reflecting their potential roles in plant growth, development or stress responses. Furthermore, we demonstrated that silencing of StKPNβ3a, a SA- and H2O2-inducible KPNβ genes led to increased susceptibility to environmental challenges, implying its crucial roles in plant adaption to abiotic stresses. Overall, our results provide molecular insights into StKPNβ gene family, which will serve as a strong foundation for further functional characterization and will facilitate potato breeding programs.
Keywords: abiotic stress; expression analysis; karyopherin; solanum tuberosum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- 2017YFD0200708/National Key Research and Development Program of China
- YCXTD0000210/Young Talent Supporting Program" of the Beijing Agricultural and Forestry Sciences
- 2018BBF02021/Key Research and Development Program of Ningxia Autonomous Region in China
- 31601622/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

