TLR4-Targeting Therapeutics: Structural Basis and Computer-Aided Drug Discovery Approaches
- PMID: 32023919
- PMCID: PMC7037830
- DOI: 10.3390/molecules25030627
TLR4-Targeting Therapeutics: Structural Basis and Computer-Aided Drug Discovery Approaches
Abstract
The integration of computational techniques into drug development has led to a substantial increase in the knowledge of structural, chemical, and biological data. These techniques are useful for handling the big data generated by empirical and clinical studies. Over the last few years, computer-aided drug discovery methods such as virtual screening, pharmacophore modeling, quantitative structure-activity relationship analysis, and molecular docking have been employed by pharmaceutical companies and academic researchers for the development of pharmacologically active drugs. Toll-like receptors (TLRs) play a vital role in various inflammatory, autoimmune, and neurodegenerative disorders such as sepsis, rheumatoid arthritis, inflammatory bowel disease, Alzheimer's disease, multiple sclerosis, cancer, and systemic lupus erythematosus. TLRs, particularly TLR4, have been identified as potential drug targets for the treatment of these diseases, and several relevant compounds are under preclinical and clinical evaluation. This review covers the reported computational studies and techniques that have provided insights into TLR4-targeting therapeutics. Furthermore, this article provides an overview of the computational methods that can benefit a broad audience in this field and help with the development of novel drugs for TLR-related disorders.
Keywords: TLR4; agonist; antagonist; computer-aided drug discovery; molecular dynamics; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

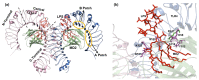

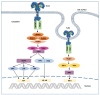
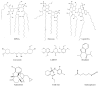
References
-
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H., Wang H., et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

