The First Non-LRV RNA Virus in Leishmania
- PMID: 32024293
- PMCID: PMC7077295
- DOI: 10.3390/v12020168
The First Non-LRV RNA Virus in Leishmania
Abstract
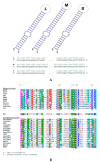
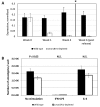
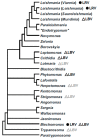
In this work, we describe the first Leishmania-infecting leishbunyavirus-the first virus other than Leishmania RNA virus (LRV) found in trypanosomatid parasites. Its host is Leishmaniamartiniquensis, a human pathogen causing infections with a wide range of manifestations from asymptomatic to severe visceral disease. This virus (LmarLBV1) possesses many characteristic features of leishbunyaviruses, such as tripartite organization of its RNA genome, with ORFs encoding RNA-dependent RNA polymerase, surface glycoprotein, and nucleoprotein on L, M, and S segments, respectively. Our phylogenetic analyses suggest that LmarLBV1 originated from leishbunyaviruses of monoxenous trypanosomatids and, probably, is a result of genomic re-assortment. The LmarLBV1 facilitates parasites' infectivity in vitro in primary murine macrophages model. The discovery of a virus in L.martiniquensis poses the question of whether it influences pathogenicity of this parasite in vivo, similarly to the LRV in other Leishmania species.
Keywords: Bunyavirales; Leishmania martiniquensis; leishbunyavirus.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

