Tracking Adeno-Associated Virus Capsid Evolution by High-Throughput Sequencing
- PMID: 32024384
- PMCID: PMC7232707
- DOI: 10.1089/hum.2019.339
Tracking Adeno-Associated Virus Capsid Evolution by High-Throughput Sequencing
Abstract
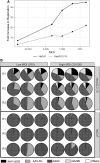
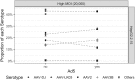
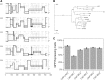
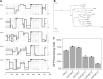
Despite early successes using recombinant adeno-associated virus (rAAV) vectors in clinical gene therapy trials, limitations remain making additional advancements a necessity. Some of the challenges include variable levels of pre-existing neutralizing antibodies and poor transduction in specific target tissues and/or diseases. In addition, readministration of an rAAV vector is in general not possible due to the immune response against the capsid. Recombinant adeno-associated virus (AAV) vectors with novel capsids can be isolated in nature or developed through different directed evolution strategies. However, in most cases, the process of AAV selection is not well understood and new strategies are required to define the best parameters to develop more efficient and functional rAAV capsids. Therefore, the use of barcoding for AAV capsid libraries, which can be screened by high-throughput sequencing, provides a powerful tool to track AAV capsid evolution and potentially improve AAV capsid library screens. In this study, we examined how different parameters affect the screen of two different AAV libraries in two human cell types. We uncovered new and unexpected insights in how to maximize the likelihood of obtaining AAV variants with the desired properties. The major findings of the study are the following. (1) Inclusion of helper-virus for AAV replication can selectively propagate variants that can replicate to higher titers, but are not necessarily better at transduction. (2) Competition between AAVs with specific capsids can take place in cells that have been infected with different AAVs. (3) The use of low multiplicity of infections for infection results in more variation between screens and is not optimal at selecting the most desired capsids. (4) Using multiple rounds of selection can be counterproductive. We conclude that each of these parameters should be taken into consideration when screening AAV libraries for enhanced properties of interest.
Keywords: AAV libraries; capsid evolution; high-throughput sequencing.
Conflict of interest statement
M.A.K. has commercial affiliations and stock and/or equity in companies with technology broadly related to this article. All other authors declare no conflicts of interest.
Figures


References
-
- Munch RC, Muth A, Muik A, et al. . Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat Commun 2015;6:6246. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

