Genomic footprints of activated telomere maintenance mechanisms in cancer
- PMID: 32024817
- PMCID: PMC7002710
- DOI: 10.1038/s41467-019-13824-9
Genomic footprints of activated telomere maintenance mechanisms in cancer
Erratum in
-
Author Correction: Genomic footprints of activated telomere maintenance mechanisms in cancer.Nat Commun. 2022 Dec 8;13(1):7574. doi: 10.1038/s41467-022-32328-7. Nat Commun. 2022. PMID: 36481818 Free PMC article. No abstract available.
Abstract
Cancers require telomere maintenance mechanisms for unlimited replicative potential. They achieve this through TERT activation or alternative telomere lengthening associated with ATRX or DAXX loss. Here, as part of the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium, we dissect whole-genome sequencing data of over 2500 matched tumor-control samples from 36 different tumor types aggregated within the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium to characterize the genomic footprints of these mechanisms. While the telomere content of tumors with ATRX or DAXX mutations (ATRX/DAXXtrunc) is increased, tumors with TERT modifications show a moderate decrease of telomere content. One quarter of all tumor samples contain somatic integrations of telomeric sequences into non-telomeric DNA. This fraction is increased to 80% prevalence in ATRX/DAXXtrunc tumors, which carry an aberrant telomere variant repeat (TVR) distribution as another genomic marker. The latter feature includes enrichment or depletion of the previously undescribed singleton TVRs TTCGGG and TTTGGG, respectively. Our systematic analysis provides new insight into the recurrent genomic alterations associated with telomere maintenance mechanisms in cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures
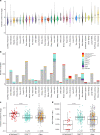



Comment in
-
Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX.Nat Commun. 2021 Mar 10;12(1):1552. doi: 10.1038/s41467-021-21794-0. Nat Commun. 2021. PMID: 33692341 Free PMC article. No abstract available.
-
Formal reply to "Alternative lengthening of telomeres is not synonymous with mutations in ATRX/DAXX".Nat Commun. 2021 Mar 10;12(1):1551. doi: 10.1038/s41467-021-21796-y. Nat Commun. 2021. PMID: 33692348 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

