High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations
- PMID: 32024823
- PMCID: PMC7002524
- DOI: 10.1038/s41467-019-13885-w
High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations
Erratum in
-
Author Correction: High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations.Nat Commun. 2022 Dec 8;13(1):7572. doi: 10.1038/s41467-022-32333-w. Nat Commun. 2022. PMID: 36481652 Free PMC article. No abstract available.
Abstract
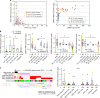
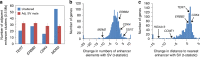
The impact of somatic structural variants (SVs) on gene expression in cancer is largely unknown. Here, as part of the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium, which aggregated whole-genome sequencing data and RNA sequencing from a common set of 1220 cancer cases, we report hundreds of genes for which the presence within 100 kb of an SV breakpoint associates with altered expression. For the majority of these genes, expression increases rather than decreases with corresponding breakpoint events. Up-regulated cancer-associated genes impacted by this phenomenon include TERT, MDM2, CDK4, ERBB2, CD274, PDCD1LG2, and IGF2. TERT-associated breakpoints involve ~3% of cases, most frequently in liver biliary, melanoma, sarcoma, stomach, and kidney cancers. SVs associated with up-regulation of PD1 and PDL1 genes involve ~1% of non-amplified cases. For many genes, SVs are significantly associated with increased numbers or greater proximity of enhancer regulatory elements near the gene. DNA methylation near the promoter is often increased with nearby SV breakpoint, which may involve inactivation of repressor elements.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

