Identification and characterization of novel filament-forming proteins in cyanobacteria
- PMID: 32024928
- PMCID: PMC7002697
- DOI: 10.1038/s41598-020-58726-9
Identification and characterization of novel filament-forming proteins in cyanobacteria
Abstract
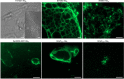
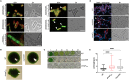
Filament-forming proteins in bacteria function in stabilization and localization of proteinaceous complexes and replicons; hence they are instrumental for myriad cellular processes such as cell division and growth. Here we present two novel filament-forming proteins in cyanobacteria. Surveying cyanobacterial genomes for coiled-coil-rich proteins (CCRPs) that are predicted as putative filament-forming proteins, we observed a higher proportion of CCRPs in filamentous cyanobacteria in comparison to unicellular cyanobacteria. Using our predictions, we identified nine protein families with putative intermediate filament (IF) properties. Polymerization assays revealed four proteins that formed polymers in vitro and three proteins that formed polymers in vivo. Fm7001 from Fischerella muscicola PCC 7414 polymerized in vitro and formed filaments in vivo in several organisms. Additionally, we identified a tetratricopeptide repeat protein - All4981 - in Anabaena sp. PCC 7120 that polymerized into filaments in vitro and in vivo. All4981 interacts with known cytoskeletal proteins and is indispensable for Anabaena viability. Although it did not form filaments in vitro, Syc2039 from Synechococcus elongatus PCC 7942 assembled into filaments in vivo and a Δsyc2039 mutant was characterized by an impaired cytokinesis. Our results expand the repertoire of known prokaryotic filament-forming CCRPs and demonstrate that cyanobacterial CCRPs are involved in cell morphology, motility, cytokinesis and colony integrity.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Rippka R, Stanier RY, Deruelles J, Herdman M, Waterbury JB. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

