Identification of a parasitic symbiosis between respiratory metabolisms in the biogeochemical chlorine cycle
- PMID: 32024948
- PMCID: PMC7174294
- DOI: 10.1038/s41396-020-0599-1
Identification of a parasitic symbiosis between respiratory metabolisms in the biogeochemical chlorine cycle
Abstract
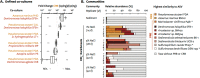
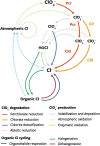
A key step in the chlorine cycle is the reduction of perchlorate (ClO4-) and chlorate (ClO3-) to chloride by microbial respiratory pathways. Perchlorate-reducing bacteria and chlorate-reducing bacteria differ in that the latter cannot use perchlorate, the most oxidized chlorine compound. However, a recent study identified a bacterium with the chlorate reduction pathway dominating a community provided only perchlorate. Here we confirm a metabolic interaction between perchlorate- and chlorate-reducing bacteria and define its mechanism. Perchlorate-reducing bacteria supported the growth of chlorate-reducing bacteria to up to 90% of total cells in communities and co-cultures. Chlorate-reducing bacteria required the gene for chlorate reductase to grow in co-culture with perchlorate-reducing bacteria, demonstrating that chlorate is responsible for the interaction, not the subsequent intermediates chlorite and oxygen. Modeling of the interaction suggested that cells specialized for chlorate reduction have a competitive advantage for consuming chlorate produced from perchlorate, especially at high concentrations of perchlorate, because perchlorate and chlorate compete for a single enzyme in perchlorate-reducing cells. We conclude that perchlorate-reducing bacteria inadvertently support large populations of chlorate-reducing bacteria in a parasitic relationship through the release of the intermediate chlorate. An implication of these findings is that undetected chlorate-reducing bacteria have likely negatively impacted efforts to bioremediate perchlorate pollution for decades.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Liebensteiner MG, Oosterkamp MJ, Stams AJM. Microbial respiration with chlorine oxyanions: diversity and physiological and biochemical properties of chlorate- and perchlorate-reducing microorganisms. Ann NY Acad Sci. 2016;1365:59–72. - PubMed
-
- Palmisano A, Hazen T. Bioremediation of metals and radionuclides: what it is and how it works, 2 edn. United States: N. 2003. p. 2003.
-
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–86. - PubMed
-
- Youngblut MD, Wang O, Barnum TP, Coates JD. (Per)chlorate in biology on earth and beyond. Annu Rev Microbiol. 2016;70:435–459. - PubMed

