Comprehensive molecular characterization of mitochondrial genomes in human cancers
- PMID: 32024997
- PMCID: PMC7058535
- DOI: 10.1038/s41588-019-0557-x
Comprehensive molecular characterization of mitochondrial genomes in human cancers
Erratum in
-
Publisher Correction: Comprehensive molecular characterization of mitochondrial genomes in human cancers.Nat Genet. 2023 May;55(5):893. doi: 10.1038/s41588-020-0587-4. Nat Genet. 2023. PMID: 32047324 Free PMC article. No abstract available.
-
Author Correction: Comprehensive molecular characterization of mitochondrial genomes in human cancers.Nat Genet. 2023 May;55(5):892. doi: 10.1038/s41588-020-0629-y. Nat Genet. 2023. PMID: 32341528 Free PMC article. No abstract available.
-
Author Correction: Comprehensive molecular characterization of mitochondrial genomes in human cancers.Nat Genet. 2023 Jun;55(6):1078. doi: 10.1038/s41588-023-01317-x. Nat Genet. 2023. PMID: 36944732 Free PMC article. No abstract available.
Abstract
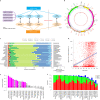
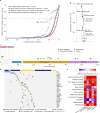
Mitochondria are essential cellular organelles that play critical roles in cancer. Here, as part of the International Cancer Genome Consortium/The Cancer Genome Atlas Pan-Cancer Analysis of Whole Genomes Consortium, which aggregated whole-genome sequencing data from 2,658 cancers across 38 tumor types, we performed a multidimensional, integrated characterization of mitochondrial genomes and related RNA sequencing data. Our analysis presents the most definitive mutational landscape of mitochondrial genomes and identifies several hypermutated cases. Truncating mutations are markedly enriched in kidney, colorectal and thyroid cancers, suggesting oncogenic effects with the activation of signaling pathways. We find frequent somatic nuclear transfers of mitochondrial DNA, some of which disrupt therapeutic target genes. Mitochondrial copy number varies greatly within and across cancers and correlates with clinical variables. Co-expression analysis highlights the function of mitochondrial genes in oxidative phosphorylation, DNA repair and the cell cycle, and shows their connections with clinically actionable genes. Our study lays a foundation for translating mitochondrial biology into clinical applications.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

