Live-animal imaging of native haematopoietic stem and progenitor cells
- PMID: 32025033
- PMCID: PMC7021587
- DOI: 10.1038/s41586-020-1971-z
Live-animal imaging of native haematopoietic stem and progenitor cells
Abstract
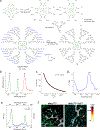
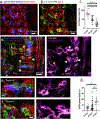
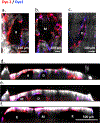
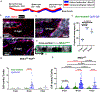
The biology of haematopoietic stem cells (HSCs) has predominantly been studied under transplantation conditions1,2. It has been particularly challenging to study dynamic HSC behaviour, given that the visualization of HSCs in the native niche in live animals has not, to our knowledge, been achieved. Here we describe a dual genetic strategy in mice that restricts reporter labelling to a subset of the most quiescent long-term HSCs (LT-HSCs) and that is compatible with current intravital imaging approaches in the calvarial bone marrow3-5. We show that this subset of LT-HSCs resides close to both sinusoidal blood vessels and the endosteal surface. By contrast, multipotent progenitor cells (MPPs) show greater variation in distance from the endosteum and are more likely to be associated with transition zone vessels. LT-HSCs are not found in bone marrow niches with the deepest hypoxia and instead are found in hypoxic environments similar to those of MPPs. In vivo time-lapse imaging revealed that LT-HSCs at steady-state show limited motility. Activated LT-HSCs show heterogeneous responses, with some cells becoming highly motile and a fraction of HSCs expanding clonally within spatially restricted domains. These domains have defined characteristics, as HSC expansion is found almost exclusively in a subset of bone marrow cavities with bone-remodelling activity. By contrast, cavities with low bone-resorbing activity do not harbour expanding HSCs. These findings point to previously unknown heterogeneity within the bone marrow microenvironment, imposed by the stages of bone turnover. Our approach enables the direct visualization of HSC behaviours and dissection of heterogeneity in HSC niches.
Conflict of interest statement
Competing interests
The authors declare no competing financial interests.
Figures











Comment in
-
Should I Stay or Should I Flow: HSCs Are on the Move!Cell Stem Cell. 2020 Aug 6;27(2):189-190. doi: 10.1016/j.stem.2020.07.014. Cell Stem Cell. 2020. PMID: 32763177
References
METHODS REFERENCES
-
- Kalajzic Z et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone 31, 654–660 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

