Altered Expression of Long Noncoding and Messenger RNAs in Diabetic Nephropathy following Treatment with Rosiglitazone
- PMID: 32025515
- PMCID: PMC6983290
- DOI: 10.1155/2020/1360843
Altered Expression of Long Noncoding and Messenger RNAs in Diabetic Nephropathy following Treatment with Rosiglitazone
Abstract
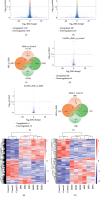
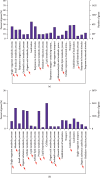
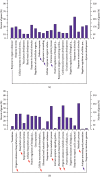
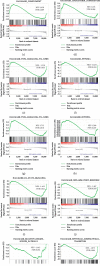
Diabetic nephropathy (DN) is characterized by metabolic disorder and inflammation. However, the regulatory effects that long noncoding RNAs (lncRNAs) have on the pathogenesis of DN and on the efficacy of rosiglitazone treatment have yet to be clearly defined. Herein, we performed unbiased RNA sequencing to characterize the transcriptomic profiles in db/db diabetic mouse model with or without rosiglitazone treatment that served to improve the phenotypes of DN. Moreover, RNA-seq profiling revealed that the development of DN caused an upregulation in the expression of 1176 mRNAs and a downregulation in the expression of 1010 mRNAs compared to controls, with the expression of 251 mRNAs being returned to normal following treatment with rosiglitazone. Further, 88 upregulated and 68 downregulated lncRNAs were identified in db/db mice compared to controls, 10 of which had their normal expression restored following treatment with rosiglitazone. Bioinformatic analysis revealed that the primary pathways involved in the pathogenesis of DN, and subsequently in the therapeutic effects of PPARγ, are related to inflammatory and metabolic processes. From bioinformatics analysis, lncRNA-AI838599 emerged as a novel molecular mechanism for rosiglitazone treatment in DN through TNFα-NFκb pathway. These findings may indicate a new molecular regulatory approach for the development of DN therapeutic agents.
Copyright © 2020 Liwen Zhang et al.
Conflict of interest statement
All authors declare that they have no conflicts of interest.
Figures


References
-
- Bichu P., Nistala R., Khan A., Sowers J. R., Whaley-Connell A. Angiotensin receptor blockers for the reduction of proteinuria in diabetic patients with overt nephropathy: results from the AMADEO study. Vascular Health and Risk Management. 2008;5(1):129–140. doi: 10.2147/vhrm.s3121. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

