DNA Sensing in the Innate Immune Response
- PMID: 32027562
- PMCID: PMC7276919
- DOI: 10.1152/physiol.00022.2019
DNA Sensing in the Innate Immune Response
Abstract
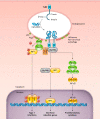
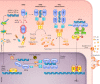
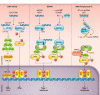
The innate immune system recognizes conserved pathogen-associated molecular patterns and produces inflammatory cytokines that direct downstream immune responses. The inappropriate localization of DNA within the cell cytosol or endosomal compartments indicates that a cell may either be infected by a DNA virus or bacterium, or has problems with its own nuclear integrity. This DNA is sensed by certain receptors that mediate cytokine production and, in some cases, initiate an inflammatory and lytic form of cell death called pyroptosis. Dysregulation of these DNA-sensing pathways is thought to contribute to autoimmune diseases and the development of cancer. In this review, we will discuss the DNA sensors Toll-like receptor 9 (TLR9), cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), absent in melanoma 2 (AIM2), and interferon gamma-inducible 16 (IFI16), their ligands, and their physiological significance. We will also examine the less-well-understood DEAH- and DEAD-box helicases DHX9, DHX36, DDX41, and RNA polymerase III, each of which may play an important role in DNA-mediated innate immunity.
Keywords: DNA sensing; autophagy; cGAS; cell death; infection; inflammasome; innate immunity; interferon.
Figures

References
-
- Algaba-Chueca F, de-Madaria E, Lozano-Ruiz B, Martínez-Cardona C, Quesada-Vázquez N, Bachiller V, Tarín F, Such J, Francés R, Zapater P, González-Navajas JM. The expression and activation of the AIM2 inflammasome correlates with inflammation and disease severity in patients with acute pancreatitis. Pancreatology 17: 364–371, 2017. doi: 10.1016/j.pan.2017.03.006. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

