BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo
- PMID: 32027622
- PMCID: PMC7191002
- DOI: 10.1172/JCI132374
BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo
Abstract
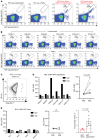
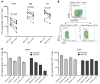
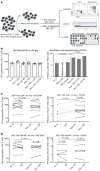
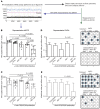
Curing HIV infection will require the elimination of a reservoir of infected CD4+ T cells that persists despite HIV-specific cytotoxic T cell (CTL) responses. Although viral latency is a critical factor in this persistence, recent evidence also suggests a role for intrinsic resistance of reservoir-harboring cells to CTL killing. This resistance may have contributed to negative outcomes of clinical trials, where pharmacologic latency reversal has thus far failed to drive reductions in HIV reservoirs. Through transcriptional profiling, we herein identified overexpression of the prosurvival factor B cell lymphoma 2 (BCL-2) as a distinguishing feature of CD4+ T cells that survived CTL killing. We show that the inducible HIV reservoir was disproportionately present in BCL-2hi subsets in ex vivo CD4+ T cells. Treatment with the BCL-2 antagonist ABT-199 was not sufficient to drive reductions in ex vivo viral reservoirs when tested either alone or with a latency-reversing agent (LRA). However, the triple combination of strong LRAs, HIV-specific T cells, and a BCL-2 antagonist uniquely enabled the depletion of ex vivo viral reservoirs. Our results provide rationale for novel therapeutic approaches targeting HIV cure and, more generally, suggest consideration of BCL-2 antagonism as a means of enhancing CTL immunotherapy in other settings, such as cancer.
Keywords: AIDS/HIV; Adaptive immunity; Cellular immune response; Immunology; T cells.
Conflict of interest statement
Figures



References
-
- Turk G, et al. Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8+ T-cell antiviral activity and correlates with preservation of the CD4+ T-cell compartment. J Virol. 2013;87(13):7445–7462. doi: 10.1128/JVI.00865-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

