Duckweed hosts a taxonomically similar bacterial assemblage as the terrestrial leaf microbiome
- PMID: 32027711
- PMCID: PMC7004381
- DOI: 10.1371/journal.pone.0228560
Duckweed hosts a taxonomically similar bacterial assemblage as the terrestrial leaf microbiome
Abstract
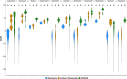
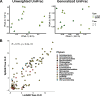
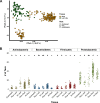
Culture-independent characterization of microbial communities associated with popular plant model systems have increased our understanding of the plant microbiome. However, the integration of other model systems, such as duckweed, could facilitate our understanding of plant microbiota assembly and evolution. Duckweeds are floating aquatic plants with many characteristics, including small size and reduced plant architecture, that suggest their use as a facile model system for plant microbiome studies. Here, we investigated the structure and assembly of the duckweed bacterial microbiome. First, a culture-independent survey of the duckweed bacterial microbiome from different locations in New Jersey revealed similar phylogenetic profiles. These studies showed that Proteobacteria is a dominant phylum in the duckweed bacterial microbiome. To observe the assembly dynamics of the duckweed bacterial community, we inoculated quasi-gnotobiotic duckweed with wastewater effluent from a municipal wastewater treatment plant. Our results revealed that duckweed strongly shapes its bacterial microbiome and forms distinct associations with bacterial community members from the initial inoculum. Additionally, these inoculation studies showed the bacterial communities of different duckweed species were similar in taxa composition and abundance. Analysis across the different duckweed bacterial communities collected in this study identified a set of "core" bacterial taxa consistently present on duckweed irrespective of the locale and context. Furthermore, comparison of the duckweed bacterial community to that of rice and Arabidopsis revealed a conserved taxonomic structure between the duckweed microbiome and the terrestrial leaf microbiome. Our results suggest that duckweeds utilize similar bacterial community assembly principles as those found in terrestrial plants and indicate a highly conserved structuring effect of leaf tissue on the plant microbiome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Massart S, Martinez-Medina M, Jijakli MH. Biological control in the microbiome era: challenges and opportunities. Biological control. 2015. October 1;89:98–108.
-
- Hacquard S, Garrido-Oter R, González A, Spaepen S, Ackermann G, Lebeis S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell host & microbe. 2015. May 13;17(5):603–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

