TNF receptor-associated factor 6 interacts with ALS-linked misfolded superoxide dismutase 1 and promotes aggregation
- PMID: 32029478
- PMCID: PMC7086032
- DOI: 10.1074/jbc.RA119.011215
TNF receptor-associated factor 6 interacts with ALS-linked misfolded superoxide dismutase 1 and promotes aggregation
Abstract
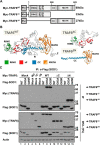
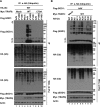
Amyotrophic lateral sclerosis (ALS) is a fatal disease, characterized by the selective loss of motor neurons leading to paralysis. Mutations in the gene encoding superoxide dismutase 1 (SOD1) are the second most common cause of familial ALS, and considerable evidence suggests that these mutations result in an increase in toxicity due to protein misfolding. We previously demonstrated in the SOD1G93A rat model that misfolded SOD1 exists as distinct conformers and forms deposits on mitochondrial subpopulations. Here, using SOD1G93A rats and conformation-restricted antibodies specific for misfolded SOD1 (B8H10 and AMF7-63), we identified the interactomes of the mitochondrial pools of misfolded SOD1. This strategy identified binding proteins that uniquely interacted with either AMF7-63 or B8H10-reactive SOD1 conformers as well as a high proportion of interactors common to both conformers. Of this latter set, we identified the E3 ubiquitin ligase TNF receptor-associated factor 6 (TRAF6) as a SOD1 interactor, and we determined that exposure of the SOD1 functional loops facilitates this interaction. Of note, this conformational change was not universally fulfilled by all SOD1 variants and differentiated TRAF6 interacting from TRAF6 noninteracting SOD1 variants. Functionally, TRAF6 stimulated polyubiquitination and aggregation of the interacting SOD1 variants. TRAF6 E3 ubiquitin ligase activity was required for the former but was dispensable for the latter, indicating that TRAF6-mediated polyubiquitination and aggregation of the SOD1 variants are independent events. We propose that the interaction between misfolded SOD1 and TRAF6 may be relevant to the etiology of ALS.
Keywords: TNF receptor–associated factor (TRAF); amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease); mitochondria; protein aggregation; protein misfolding; superoxide dismutase (SOD); ubiquitin.
© 2020 Semmler et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

