Alternative polyadenylation drives oncogenic gene expression in pancreatic ductal adenocarcinoma
- PMID: 32029502
- PMCID: PMC7111527
- DOI: 10.1101/gr.257550.119
Alternative polyadenylation drives oncogenic gene expression in pancreatic ductal adenocarcinoma
Abstract
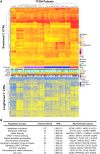
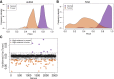
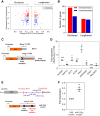
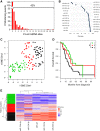
Alternative polyadenylation (APA) is a gene regulatory process that dictates mRNA 3'-UTR length, resulting in changes in mRNA stability and localization. APA is frequently disrupted in cancer and promotes tumorigenesis through altered expression of oncogenes and tumor suppressors. Pan-cancer analyses have revealed common APA events across the tumor landscape; however, little is known about tumor type-specific alterations that may uncover novel events and vulnerabilities. Here, we integrate RNA-sequencing data from the Genotype-Tissue Expression (GTEx) project and The Cancer Genome Atlas (TCGA) to comprehensively analyze APA events in 148 pancreatic ductal adenocarcinomas (PDACs). We report widespread, recurrent, and functionally relevant 3'-UTR alterations associated with gene expression changes of known and newly identified PDAC growth-promoting genes and experimentally validate the effects of these APA events on protein expression. We find enrichment for APA events in genes associated with known PDAC pathways, loss of tumor-suppressive miRNA binding sites, and increased heterogeneity in 3'-UTR forms of metabolic genes. Survival analyses reveal a subset of 3'-UTR alterations that independently characterize a poor prognostic cohort among PDAC patients. Finally, we identify and validate the casein kinase CSNK1A1 (also known as CK1alpha or CK1a) as an APA-regulated therapeutic target in PDAC. Knockdown or pharmacological inhibition of CSNK1A1 attenuates PDAC cell proliferation and clonogenic growth. Our single-cancer analysis reveals APA as an underappreciated driver of protumorigenic gene expression in PDAC via the loss of miRNA regulation.
© 2020 Venkat et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
