A novel NanoBiT-based assay monitors the interaction between lipoprotein lipase and GPIHBP1 in real time
- PMID: 32029511
- PMCID: PMC7112140
- DOI: 10.1194/jlr.D119000388
A novel NanoBiT-based assay monitors the interaction between lipoprotein lipase and GPIHBP1 in real time
Abstract
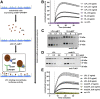
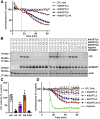
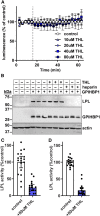
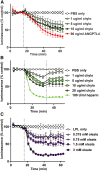
The hydrolysis of triglycerides in triglyceride-rich lipoproteins by LPL is critical for the delivery of triglyceride-derived fatty acids to tissues, including heart, skeletal muscle, and adipose tissues. Physiologically active LPL is normally bound to the endothelial cell protein glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1 (GPIHBP1), which transports LPL across endothelial cells, anchors LPL to the vascular wall, and stabilizes LPL activity. Disruption of LPL-GPIHBP1 binding significantly alters triglyceride metabolism and lipid partitioning. In this study, we modified the NanoLuc® Binary Technology split-luciferase system to develop a novel assay that monitors the binding of LPL to GPIHBP1 on endothelial cells in real time. We validated the specificity and sensitivity of the assay using endothelial lipase and a mutant version of LPL and found that this assay reliably and specifically detected the interaction between LPL and GPIHBP1. We then interrogated various endogenous and exogenous inhibitors of LPL-mediated lipolysis for their ability to disrupt the binding of LPL to GPIHBP1. We found that angiopoietin-like (ANGPTL)4 and ANGPTL3-ANGPTL8 complexes disrupted the interactions of LPL and GPIHBP1, whereas the exogenous LPL blockers we tested (tyloxapol, poloxamer-407, and tetrahydrolipstatin) did not. We also found that chylomicrons could dissociate LPL from GPIHBP1 and found evidence that this dissociation was mediated in part by the fatty acids produced by lipolysis. These results demonstrate the ability of this assay to monitor LPL-GPIHBP1 binding and to probe how various agents influence this important complex.
Keywords: NanoLuc® Binary Technology; chylomicrons; endothelial cell; glycosylphosphatidylinositol-anchored high density lipoprotein binding protein 1; lipolysis; lipoprotein metabolism; triglyceride.
Copyright © 2020 Shetty et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Angiopoietin-like 4 Modifies the Interactions between Lipoprotein Lipase and Its Endothelial Cell Transporter GPIHBP1.J Biol Chem. 2015 May 8;290(19):11865-77. doi: 10.1074/jbc.M114.623769. Epub 2015 Mar 25. J Biol Chem. 2015. PMID: 25809481 Free PMC article.
-
ANGPTL8 promotes the ability of ANGPTL3 to bind and inhibit lipoprotein lipase.Mol Metab. 2017 Oct;6(10):1137-1149. doi: 10.1016/j.molmet.2017.06.014. Epub 2017 Jun 29. Mol Metab. 2017. PMID: 29031715 Free PMC article.
-
Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons.Cell Metab. 2007 Apr;5(4):279-91. doi: 10.1016/j.cmet.2007.02.002. Cell Metab. 2007. PMID: 17403372 Free PMC article.
-
Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 and the intravascular processing of triglyceride-rich lipoproteins.J Intern Med. 2012 Dec;272(6):528-40. doi: 10.1111/joim.12003. Epub 2012 Nov 1. J Intern Med. 2012. PMID: 23020258 Free PMC article. Review.
-
GPIHBP1, an endothelial cell transporter for lipoprotein lipase.J Lipid Res. 2011 Nov;52(11):1869-84. doi: 10.1194/jlr.R018689. Epub 2011 Aug 15. J Lipid Res. 2011. PMID: 21844202 Free PMC article. Review.
Cited by
-
The response to fasting and refeeding reveals functional regulation of lipoprotein lipase proteoforms.Front Physiol. 2023 Oct 16;14:1271149. doi: 10.3389/fphys.2023.1271149. eCollection 2023. Front Physiol. 2023. PMID: 37916217 Free PMC article.
-
Lipoprotein Lipase: Structure, Function, and Genetic Variation.Genes (Basel). 2025 Jan 5;16(1):55. doi: 10.3390/genes16010055. Genes (Basel). 2025. PMID: 39858602 Free PMC article. Review.
-
Structure of dimeric lipoprotein lipase reveals a pore adjacent to the active site.Nat Commun. 2023 May 4;14(1):2569. doi: 10.1038/s41467-023-38243-9. Nat Commun. 2023. PMID: 37142573 Free PMC article.
-
The intrinsic instability of the hydrolase domain of lipoprotein lipase facilitates its inactivation by ANGPTL4-catalyzed unfolding.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2026650118. doi: 10.1073/pnas.2026650118. Proc Natl Acad Sci U S A. 2021. PMID: 33723082 Free PMC article.
-
Angiopoietin-Like Protein 3 (ANGPTL3) Modulates Lipoprotein Metabolism and Dyslipidemia.Int J Mol Sci. 2021 Jul 7;22(14):7310. doi: 10.3390/ijms22147310. Int J Mol Sci. 2021. PMID: 34298929 Free PMC article. Review.
References
-
- Beigneux A. P., Davies B. S. J., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., et al. . 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. - PMC - PubMed
-
- Kristensen K. K., Midtgaard S. R., Mysling S., Kovrov O., Hansen L. B., Skar-Gislinge N., Beigneux A. P., Kragelund B. B., Olivecrona G., Young S. G., et al. . 2018. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 115: E6020–E6029. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

