CRISPR-engineered T cells in patients with refractory cancer
- PMID: 32029687
- PMCID: PMC11249135
- DOI: 10.1126/science.aba7365
CRISPR-engineered T cells in patients with refractory cancer
Abstract
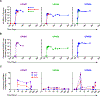
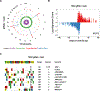
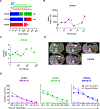
CRISPR-Cas9 gene editing provides a powerful tool to enhance the natural ability of human T cells to fight cancer. We report a first-in-human phase 1 clinical trial to test the safety and feasibility of multiplex CRISPR-Cas9 editing to engineer T cells in three patients with refractory cancer. Two genes encoding the endogenous T cell receptor (TCR) chains, TCRα (TRAC) and TCRβ (TRBC), were deleted in T cells to reduce TCR mispairing and to enhance the expression of a synthetic, cancer-specific TCR transgene (NY-ESO-1). Removal of a third gene encoding programmed cell death protein 1 (PD-1; PDCD1), was performed to improve antitumor immunity. Adoptive transfer of engineered T cells into patients resulted in durable engraftment with edits at all three genomic loci. Although chromosomal translocations were detected, the frequency decreased over time. Modified T cells persisted for up to 9 months, suggesting that immunogenicity is minimal under these conditions and demonstrating the feasibility of CRISPR gene editing for cancer immunotherapy.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures




Comment in
-
Knocking out barriers to engineered cell activity.Science. 2020 Feb 28;367(6481):976-977. doi: 10.1126/science.aba9844. Epub 2020 Feb 6. Science. 2020. PMID: 32029685 No abstract available.
-
CRISPR-Edited Immune Effectors: The End of the Beginning.Mol Ther. 2020 Apr 8;28(4):995-996. doi: 10.1016/j.ymthe.2020.03.009. Epub 2020 Mar 22. Mol Ther. 2020. PMID: 32203679 Free PMC article. No abstract available.
-
A CRISPR Odyssey into Cancer Immunotherapy.CRISPR J. 2020 Apr;3(2):73-75. doi: 10.1089/crispr.2020.29090.ede. CRISPR J. 2020. PMID: 32315222 Free PMC article. No abstract available.
-
Oncology Scan: Radiation Biology and Genomic Predictors of Response.Int J Radiat Oncol Biol Phys. 2020 Jul 1;107(3):393-397. doi: 10.1016/j.ijrobp.2020.04.008. Int J Radiat Oncol Biol Phys. 2020. PMID: 32531379 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

