Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions
- PMID: 32029752
- PMCID: PMC7005164
- DOI: 10.1038/s41598-020-58563-w
Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions
Abstract
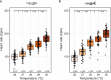
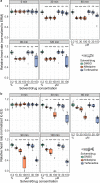
Accurate quantification of heartbeats in fish models is an important readout to study cardiovascular biology, disease states and pharmacology. However, dependence on anaesthesia, laborious sample orientation or requirement for fluorescent reporters have hampered the use of high-throughput heartbeat analysis. To overcome these limitations, we established an efficient screening assay employing automated label-free heart rate determination of randomly oriented, non-anesthetized medaka (Oryzias latipes) and zebrafish (Danio rerio) embryos in microtiter plates. Automatically acquired bright-field data feeds into an easy-to-use HeartBeat software with graphical user interface for automated quantification of heart rate and rhythm. Sensitivity of the assay was demonstrated by profiling heart rates during entire embryonic development. Our analysis revealed rapid adaption of heart rates to temperature changes, which has implications for standardization of experimental layout. The assay allows scoring of multiple embryos per well enabling a throughput of >500 embryos per 96-well plate. In a proof of principle screen for compound testing, we captured concentration-dependent effects of nifedipine and terfenadine over time. Our novel assay permits large-scale applications ranging from phenotypic screening, interrogation of gene functions to cardiovascular drug development.
Conflict of interest statement
Jochen Gehrig is an employee of DITABIS AG, Pforzheim, Germany. All other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

