A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing
- PMID: 32029767
- PMCID: PMC7004982
- DOI: 10.1038/s41598-020-58410-y
A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing
Erratum in
-
Author Correction: A cross-sectional study to characterize local HIV-1 dynamics in Washington, DC using next-generation sequencing.Sci Rep. 2020 Apr 22;10(1):7099. doi: 10.1038/s41598-020-63859-y. Sci Rep. 2020. PMID: 32317740 Free PMC article.
Abstract
Washington, DC continues to experience a generalized HIV-1 epidemic. We characterized the local phylodynamics of HIV-1 in DC using next-generation sequencing (NGS) data. Viral samples from 68 participants from 2016 through 2017 were sequenced and paired with epidemiological data. Phylogenetic and network inferences, drug resistant mutations (DRMs), subtypes and HIV-1 diversity estimations were completed. Haplotypes were reconstructed to infer transmission clusters. Phylodynamic inferences based on the HIV-1 polymerase (pol) and envelope genes (env) were compared. Higher HIV-1 diversity (n.s.) was seen in men who have sex with men, heterosexual, and male participants in DC. 54.0% of the participants contained at least one DRM. The 40-49 year-olds showed the highest prevalence of DRMs (22.9%). Phylogenetic analysis of pol and env sequences grouped 31.9-33.8% of the participants into clusters. HIV-TRACE grouped 2.9-12.8% of participants when using consensus sequences and 9.0-64.2% when using haplotypes. NGS allowed us to characterize the local phylodynamics of HIV-1 in DC more broadly and accurately, given a better representation of its diversity and dynamics. Reconstructed haplotypes provided novel and deeper phylodynamic insights, which led to networks linking a higher number of participants. Our understanding of the HIV-1 epidemic was expanded with the powerful coupling of HIV-1 NGS data with epidemiological data.
Conflict of interest statement
The authors declare no competing interests.
Figures

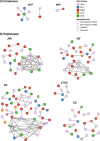
References
-
- District of Columbia Department of Health HIV/AIDS, H., STD and TB Administration (HAHSTA),. Annual Epidemiology & Surveillance Report: Surveillance Data Through December 2015. Washinton, DC Department of Health, (2016).
-
- District of Columbia Department of Health HIV/AIDS, H., STD and TB Administration (HAHSTA),. Annual Epidemiology & Surveillance Report: Surveillance Data Through December 2016. Washinton, DC Department of Health, (2017).
-
- District of Columbia Department of Health HIV/AIDS, H., STD and TB Administration (HAHSTA),. Annual Epidemiology & Surveillance Report: Surveillance Data Through December 2017. Washinton, DC Department of Health, (2018).
-
- District of Columbia Department of Health HIV/AIDS, H., STD and TB Administration (HAHSTA),. Annual Epidemiology & Surveillance Report: Surveillance Data Through December 2018. (2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

