Identifying multi-hit carcinogenic gene combinations: Scaling up a weighted set cover algorithm using compressed binary matrix representation on a GPU
- PMID: 32029803
- PMCID: PMC7005272
- DOI: 10.1038/s41598-020-58785-y
Identifying multi-hit carcinogenic gene combinations: Scaling up a weighted set cover algorithm using compressed binary matrix representation on a GPU
Abstract
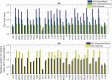
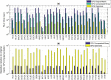
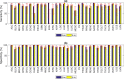
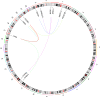
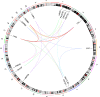
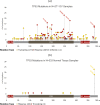
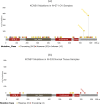
Despite decades of research, effective treatments for most cancers remain elusive. One reason is that different instances of cancer result from different combinations of multiple genetic mutations (hits). Therefore, treatments that may be effective in some cases are not effective in others. We previously developed an algorithm for identifying combinations of carcinogenic genes with mutations (multi-hit combinations), which could suggest a likely cause for individual instances of cancer. Most cancers are estimated to require three or more hits. However, the computational complexity of the algorithm scales exponentially with the number of hits, making it impractical for identifying combinations of more than two hits. To identify combinations of greater than two hits, we used a compressed binary matrix representation, and optimized the algorithm for parallel execution on an NVIDIA V100 graphics processing unit (GPU). With these enhancements, the optimized GPU implementation was on average an estimated 12,144 times faster than the original integer matrix based CPU implementation, for the 3-hit algorithm, allowing us to identify 3-hit combinations. The 3-hit combinations identified using a training set were able to differentiate between tumor and normal samples in a separate test set with 90% overall sensitivity and 93% overall specificity. We illustrate how the distribution of mutations in tumor and normal samples in the multi-hit gene combinations can suggest potential driver mutations for further investigation. With experimental validation, these combinations may provide insight into the etiology of cancer and a rational basis for targeted combination therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

