Red versus green leaves: transcriptomic comparison of foliar senescence between two Prunus cerasifera genotypes
- PMID: 32029804
- PMCID: PMC7005320
- DOI: 10.1038/s41598-020-58878-8
Red versus green leaves: transcriptomic comparison of foliar senescence between two Prunus cerasifera genotypes
Abstract
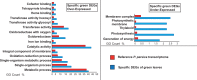
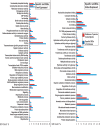
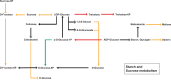
The final stage of leaf ontogenesis is represented by senescence, a highly regulated process driven by a sequential cellular breakdown involving, as the first step, chloroplast dismantling with consequent reduction of photosynthetic efficiency. Different processes, such as pigment accumulation, could protect the vulnerable photosynthetic apparatus of senescent leaves. Although several studies have produced transcriptomic data on foliar senescence, just few works have attempted to explain differences in red and green leaves throughout ontogenesis. In this work, a transcriptomic approach was used on green and red leaves of Prunus cerasifera to unveil molecular differences from leaf maturity to senescence. Our analysis revealed a higher gene regulation in red leaves compared to green ones, during leaf transition. Most of the observed DEGs were shared and involved in transcription factor activities, senescing processes and cell wall remodelling. Significant differences were detected in cellular functions: genes related to photosystem I and II were highly down-regulated in the green genotype, whereas transcripts involved in flavonoid biosynthesis, such as UDP glucose-flavonoid-3-O-glucosyltransferase (UFGT) were exclusively up-regulated in red leaves. In addition, cellular functions involved in stress response (glutathione-S-transferase, Pathogen-Related) and sugar metabolism, such as three threalose-6-phosphate synthases, were activated in senescent red leaves. In conclusion, data suggests that P. cerasifera red genotypes can regulate a set of genes and molecular mechanisms that cope with senescence, promoting more advantages during leaf ontogenesis than compared to the green ones.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kim, H. J., Lim, P. O. & Nam, H. G. Molecular regulation of senescence in Annual Plant Reviews (ed. Gan, S.) 180–181 (2007).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

