Pharmacological screening and transcriptomic functional analyses identify a synergistic interaction between dasatinib and olaparib in triple-negative breast cancer
- PMID: 32032474
- PMCID: PMC7077558
- DOI: 10.1111/jcmm.14980
Pharmacological screening and transcriptomic functional analyses identify a synergistic interaction between dasatinib and olaparib in triple-negative breast cancer
Abstract
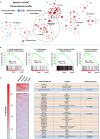
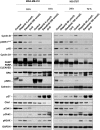
Identification of druggable vulnerabilities is a main objective in triple-negative breast cancer (TNBC), where no curative therapies exist. Gene set enrichment analyses (GSEA) and a pharmacological evaluation using a library of compounds were used to select potential druggable combinations. MTT and studies with semi-solid media were performed to explore the activity of the combinations. TNBC cell lines (MDAMB-231, BT549, HS-578T and HCC3153) and an additional panel of 16 cell lines were used to assess the activity of the two compounds. Flow cytometry experiments and biochemical studies were also performed to explore the mechanism of action. GSEA were performed using several data sets (GSE21422, GSE26910, GSE3744, GSE65194 and GSE42568), and more than 35 compounds against the identified functions were evaluated to discover druggable opportunities. Analyses done with the Chou and Talalay algorithm confirmed the synergy of dasatinib and olaparib. The combination of both agents significantly induced apoptosis in a caspase-dependent manner and revealed a pleotropic effect on cell cycle: Dasatinib arrested cells in G0/G1 and olaparib in G2/M. Dasatinib inhibited pChk1 and induced DNA damage measured by pH2AX, and olaparib increased pH3. Finally, the effect of the combination was also evaluated in a panel of 18 cell lines representative of the most frequent solid tumours, observing a particularly synergism in ovarian cancer. Breast cancer, triple negative, dasatinib, olaparib, screening.
Keywords: breast cancer; dasatinib; olaparib; screening; triple negative.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Amir E, Seruga B, Serrano R, Ocana A. Targeting DNA repair in breast cancer: a clinical and translational update. Cancer Treat Rev. 2010;36:557‐565. - PubMed
-
- Dietlein F, Thelen L, Reinhardt HC. Cancer‐specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends in genetics. 2014;30:326‐339. - PubMed
-
- Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple‐negative breast cancer‐the road to new treatment strategies. Lancet. 2016;389(10087):2430‐2442. - PubMed
-
- Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814‐819. - PubMed

