Defining the Adult Neural Stem Cell Niche Proteome Identifies Key Regulators of Adult Neurogenesis
- PMID: 32032526
- PMCID: PMC7005820
- DOI: 10.1016/j.stem.2020.01.002
Defining the Adult Neural Stem Cell Niche Proteome Identifies Key Regulators of Adult Neurogenesis
Abstract
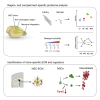
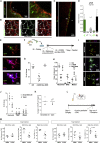
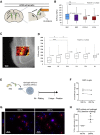
The mammalian brain contains few niches for neural stem cells (NSCs) capable of generating new neurons, whereas other regions are primarily gliogenic. Here we leverage the spatial separation of the sub-ependymal zone NSC niche and the olfactory bulb, the region to which newly generated neurons from the sub-ependymal zone migrate and integrate, and present a comprehensive proteomic characterization of these regions in comparison to the cerebral cortex, which is not conducive to neurogenesis and integration of new neurons. We find differing compositions of regulatory extracellular matrix (ECM) components in the neurogenic niche. We further show that quiescent NSCs are the main source of their local ECM, including the multi-functional enzyme transglutaminase 2, which we show is crucial for neurogenesis. Atomic force microscopy corroborated indications from the proteomic analyses that neurogenic niches are significantly stiffer than non-neurogenic parenchyma. Together these findings provide a powerful resource for unraveling unique compositions of neurogenic niches.
Keywords: C1ql3; S100a6; cerebral cortex; extracellular matrix; neuroblast; olfactory bulb; subventricular zone; tissue stiffness; transglutaminase2; transit-amplifying progenitor.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Comment in
-
Evaluation of Kjell et al.: Defining the Adult Neural Stem Cell Niche Proteome Identifies Key Regulators of Adult Neurogenesis.Cell Stem Cell. 2020 Feb 6;26(2):127-128. doi: 10.1016/j.stem.2020.01.014. Cell Stem Cell. 2020. PMID: 32032519
References
-
- Barker R.A., Götz M., Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. - PubMed
-
- Baser A., Skabkin M., Kleber S., Dang Y., Gülcüler Balta G.S., Kalamakis G., Göpferich M., Ibañez D.C., Schefzik R., Lopez A.S. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature. 2019;566:100–104. - PubMed
-
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

