HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis
- PMID: 32033571
- PMCID: PMC7006146
- DOI: 10.1186/s12981-020-0263-x
HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis
Abstract
Background: Entry inhibitors, such as Maraviroc, hold promise as components of HIV treatment and/or pre-exposure prophylaxis in Africa. Maraviroc inhibits the interaction between HIV Envelope gp120 V3-loop and CCR5 coreceptor. HIV-1 subtype C (HIV-1-C) is predominant in Southern Africa and preferably uses CCR5 co-receptor. Therefore, a significant proportion of HIV-1-C CXCR4 utilizing viruses (X4) may compromise the effectiveness of Maraviroc. This analysis examined coreceptor preferences in early and chronic HIV-1-C infections across Africa.
Methods: African HIV-1-C Envelope gp120 V3-loop sequences sampled from 1988 to 2014 were retrieved from Los Alamos HIV Sequence Database. Sequences from early infections (< 186 days post infection) and chronic infections (> 186 days post infection) were analysed for predicted co-receptor preferences using Geno2Pheno [Coreceptor] 10% FPR, Phenoseq-C, and PSSMsinsi web tools. V3-loop diversity was determined, and viral subtype was confirmed by phylogenetic analysis. National treatment guidelines across Africa were reviewed for Maraviroc recommendation.
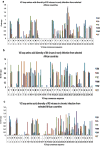
Results: Sequences from early (n = 6316) and chronic (n = 7338) HIV-1-C infected individuals from 10 and 15 African countries respectively were available for analyses. Overall, 518/6316 (8.2%; 95% CI 0.7-9.3) of early sequences were X4, with Ethiopia and Malawi having more than 10% each. For chronic infections, 8.3% (95% CI 2.4-16.2) sequences were X4 viruses, with Ethiopia, Tanzania, and Zimbabwe having more than 10% each. For sequences from early chronic infections (< 1 year post infection), the prevalence of X4 viruses was 8.5% (95% CI 2.6-11.2). In late chronic infections (≥ 5 years post infection), X4 viruses were observed in 36% (95% CI - 16.3 to 49.9), with two countries having relatively high X4 viruses: South Africa (43%) and Malawi (24%). The V3-loop amino acid sequence were more variable in X4 viruses in chronic infections compared to acute infections, with South Africa, Ethiopia and Zimbabwe showing the highest levels of V3-loop diversity. All sequences were phylogenetically confirmed as HIV-1-C and clustered according to their co-receptor tropism. In Africa, Maraviroc is registered only in South Africa and Uganda.
Conclusions: Our analyses illustrate that X4 viruses are present in significantly similar proportions in early and early chronic HIV-1 subtype C infected individuals across Africa. In contrast, in late chronic infections, X4 viruses increase 3-5 folds. We can draw two inferences from our observations: (1) to enhance the utility of Maraviroc in chronic HIV subtype C infections in Africa, prior virus co-receptor determination is needed; (2) on the flip side, research on the efficacy of CXCR4 antagonists for HIV-1-C infections is encouraged. Currently, the use of Maraviroc is very limited in Africa.
Keywords: Africa; Chronic infections; Co-receptor tropism; Early infections; HIV-1 subtype C.
Conflict of interest statement
DMT is an Associate Editor of AIDS Research and Therapy. NDM and POB declare that they have no competing interests.
Figures



Similar articles
-
Next generation sequencing reveals a high frequency of CXCR4 utilizing viruses in HIV-1 chronically infected drug experienced individuals in South Africa.J Clin Virol. 2018 Jun;103:81-87. doi: 10.1016/j.jcv.2018.02.008. Epub 2018 Feb 15. J Clin Virol. 2018. PMID: 29661652 Free PMC article.
-
Deep-Sequencing Analysis of the Dynamics of HIV-1 Quasiespecies in Naive Patients during a Short Exposure to Maraviroc.J Virol. 2018 May 14;92(11):e00390-18. doi: 10.1128/JVI.00390-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563289 Free PMC article. Clinical Trial.
-
HIV-1 coreceptor tropism in India: increasing proportion of X4-tropism in subtype C strains over two decades.J Acquir Immune Defic Syndr. 2014 Apr 1;65(4):397-404. doi: 10.1097/QAI.0000000000000046. J Acquir Immune Defic Syndr. 2014. PMID: 24189148
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
Cited by
-
Prediction of Coreceptor Tropism in HIV-1 Subtype C in Botswana.Viruses. 2023 Jan 31;15(2):403. doi: 10.3390/v15020403. Viruses. 2023. PMID: 36851617 Free PMC article.
-
HIV-1 diversity and pre-treatment drug resistance in the era of integrase inhibitor among newly diagnosed ART-naïve adult patients in Luanda, Angola.Sci Rep. 2024 Jul 10;14(1):15893. doi: 10.1038/s41598-024-66905-1. Sci Rep. 2024. PMID: 38987263 Free PMC article.
-
Unique profile of predominant CCR5-tropic in CRF07_BC HIV-1 infections and discovery of an unusual CXCR4-tropic strain.Front Immunol. 2022 Sep 23;13:911806. doi: 10.3389/fimmu.2022.911806. eCollection 2022. Front Immunol. 2022. PMID: 36211390 Free PMC article.
References
-
- UNAIDS. Fact sheet—global AIDS update 2019. UNAIDS. 2019. https://www.unaids.org/en/resources/fact-sheet. Accessed Aug 2019.
-
- Yotebieng M, Mpody C, Ravelomanana NL, Tabala M, Malongo F, Kawende B, et al. HIV viral suppression among pregnant and breastfeeding women in routine care in the Kinshasa province: a baseline evaluation of participants in CQI-PMTCT study. J Int AIDS Soc. 2019 doi: 10.1002/jia2.25376. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

