Post-mortem Plasma Cell-Free DNA Sequencing: Proof-of-Concept Study for the "Liquid Autopsy"
- PMID: 32034265
- PMCID: PMC7005783
- DOI: 10.1038/s41598-020-59193-y
Post-mortem Plasma Cell-Free DNA Sequencing: Proof-of-Concept Study for the "Liquid Autopsy"
Abstract
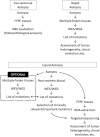
Recent genomic studies on cancer tissues obtained during rapid autopsy have provided insights into the clonal evolution and heterogeneity of cancer. However, post-mortem blood has not been subjected to genetic analyses in relation to cancer. We first confirmed that substantial quantities of cell-free DNA were present in the post-mortem plasma of 12 autopsy cases. Then, we focused on a pilot case of prostate cancer with multiple metastases for genetic analyses. Whole-exome sequencing of post-mortem plasma-derived cell-free DNA and eight frozen metastatic cancer tissues collected during rapid autopsy was performed, and compared their mutational statuses. The post-mortem plasma cell-free DNA was successfully sequenced and 344 mutations were identified. Of these, 160 were detected in at least one of the metastases. Further, 99% of the mutations shared by all metastases were present in the plasma. Sanger sequencing of 30 additional formalin-fixed metastases enabled us to map the clones harboring mutations initially detected only in the plasma. In conclusion, post-mortem blood, which is usually disposed of during conventional autopsies, can provide valuable data if sequenced in detail, especially regarding cancer heterogeneity. Furthermore, post-mortem plasma cell-free DNA sequencing (liquid autopsy) can be a novel platform for cancer research and a tool for genomic pathology.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Exome sequencing-based molecular autopsy of formalin-fixed paraffin-embedded tissue after sudden death.Genet Med. 2017 Oct;19(10):1127-1133. doi: 10.1038/gim.2017.15. Epub 2017 Mar 23. Genet Med. 2017. PMID: 28333919
-
A pilot study of next generation sequencing-liquid biopsy on cell-free DNA as a novel non-invasive diagnostic tool for Klippel-Trenaunay syndrome.Vascular. 2021 Feb;29(1):85-91. doi: 10.1177/1708538120936421. Epub 2020 Jun 26. Vascular. 2021. PMID: 32588787
-
Mutational profiles of breast cancer metastases from a rapid autopsy series reveal multiple evolutionary trajectories.JCI Insight. 2017 Dec 21;2(24):e96896. doi: 10.1172/jci.insight.96896. JCI Insight. 2017. PMID: 29263308 Free PMC article.
-
Whole exome sequencing of cell-free DNA - A systematic review and Bayesian individual patient data meta-analysis.Cancer Treat Rev. 2020 Feb;83:101951. doi: 10.1016/j.ctrv.2019.101951. Epub 2019 Dec 13. Cancer Treat Rev. 2020. PMID: 31874446
-
Dutch guideline for clinical foetal-neonatal and paediatric post-mortem radiology, including a review of literature.Eur J Pediatr. 2018 Jun;177(6):791-803. doi: 10.1007/s00431-018-3135-9. Epub 2018 Apr 19. Eur J Pediatr. 2018. PMID: 29675642 Free PMC article. Review.
Cited by
-
Reviving the Autopsy for Modern Cancer Evolution Research.Cancers (Basel). 2021 Jan 22;13(3):409. doi: 10.3390/cancers13030409. Cancers (Basel). 2021. PMID: 33499137 Free PMC article. Review.
-
Liquid biopsy for children with central nervous system tumours: Clinical integration and technical considerations.Front Pediatr. 2022 Nov 16;10:957944. doi: 10.3389/fped.2022.957944. eCollection 2022. Front Pediatr. 2022. PMID: 36467471 Free PMC article. Review.
-
Research autopsy programmes in oncology: shared experience from 14 centres across the world.J Pathol. 2024 Jun;263(2):150-165. doi: 10.1002/path.6271. Epub 2024 Mar 29. J Pathol. 2024. PMID: 38551513 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

