ZEBrA: Zebra finch Expression Brain Atlas-A resource for comparative molecular neuroanatomy and brain evolution studies
- PMID: 32037563
- PMCID: PMC8219259
- DOI: 10.1002/cne.24879
ZEBrA: Zebra finch Expression Brain Atlas-A resource for comparative molecular neuroanatomy and brain evolution studies
Abstract
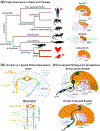
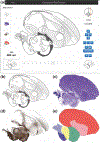
An in-depth understanding of the genetics and evolution of brain function and behavior requires a detailed mapping of gene expression in functional brain circuits across major vertebrate clades. Here we present the Zebra finch Expression Brain Atlas (ZEBrA; www.zebrafinchatlas.org, RRID: SCR_012988), a web-based resource that maps the expression of genes linked to a broad range of functions onto the brain of zebra finches. ZEBrA is a first of its kind gene expression brain atlas for a bird species and a first for any sauropsid. ZEBrA's >3,200 high-resolution digital images of in situ hybridized sections for ~650 genes (as of June 2019) are presented in alignment with an annotated histological atlas and can be browsed down to cellular resolution. An extensive relational database connects expression patterns to information about gene function, mouse expression patterns and phenotypes, and gene involvement in human diseases and communication disorders. By enabling brain-wide gene expression assessments in a bird, ZEBrA provides important substrates for comparative neuroanatomy and molecular brain evolution studies. ZEBrA also provides unique opportunities for linking genetic pathways to vocal learning and motor control circuits, as well as for novel insights into the molecular basis of sex steroids actions, brain dimorphisms, reproductive and social behaviors, sleep function, and adult neurogenesis, among many fundamental themes.
Keywords: brain atlas; brain organization; functional circuits; gene expression; molecular signature; song system; zebra finch.
© 2020 Wiley Periodicals, Inc.
Figures


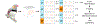











References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

