Epigenome-wide analysis of common warts reveals aberrant promoter methylation
- PMID: 32038103
- PMCID: PMC6990892
- DOI: 10.7150/ijms.39261
Epigenome-wide analysis of common warts reveals aberrant promoter methylation
Abstract
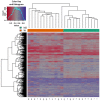
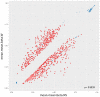
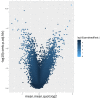
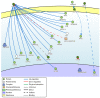
Epigenetic alteration of host DNA is a common occurrence in both low- and high-risk human papillomavirus (HPV) infection. Although changes in promoter methylation have been widely studied in HPV-associated cancers, they have not been the subject of much investigation in HPV-induced warts, which are a temporary manifestation of HPV infection. The present study sought to examine the differences in promoter methylation between warts and normal skin. To achieve this, DNA was extracted from 24 paired wart and normal skin samples and inputted into the Infinium MethylationEPIC BeadChip microarray. Differential methylation analysis revealed a clear pattern of hyper- and hypomethylation in warts compared to normal skin, and the most differentially methylated promoters were found within the EIF3EP2, CYSLTR1, C10orf99, KRT6B, LAMA4, and H3F3B genes as well as the C9orf30 pseudogene. Moreover, pathway analysis showed that the H3F3A, CDKN1A, and MAPK13 genes were the most common regulators among the most differentially methylated promoters. Since the tissue samples were excised from active warts, however, this differential methylation could either be a cellular response to HPV infection or an HPV-driven process to establish the wart and/or promote disease progression. Conclusively, it is apparent that HPV infection alters the methylation status of certain genes to possibly initiate the formation of a wart and maintain its presence.
Keywords: HPV; epigenetics; methylation; promoter; wart.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures









Similar articles
-
Genome-Wide Tiling Array Analysis of HPV-Induced Warts Reveals Aberrant Methylation of Protein-Coding and Non-Coding Regions.Genes (Basel). 2019 Dec 27;11(1):34. doi: 10.3390/genes11010034. Genes (Basel). 2019. PMID: 31892232 Free PMC article.
-
Genome-Wide CpG Island Methylation Profiles of Cutaneous Skin with and without HPV Infection.Int J Mol Sci. 2019 Sep 28;20(19):4822. doi: 10.3390/ijms20194822. Int J Mol Sci. 2019. PMID: 31569353 Free PMC article.
-
Genome-wide identification of methylated CpG sites in nongenital cutaneous warts.BMC Med Genomics. 2020 Jul 8;13(1):100. doi: 10.1186/s12920-020-00745-6. BMC Med Genomics. 2020. PMID: 32641122 Free PMC article.
-
Epigenetics, drugs of abuse, and the retroviral promoter.J Neuroimmune Pharmacol. 2013 Dec;8(5):1181-96. doi: 10.1007/s11481-013-9508-y. Epub 2013 Nov 12. J Neuroimmune Pharmacol. 2013. PMID: 24218017 Free PMC article. Review.
-
Smoking: Is it a Risk Factor for Common Warts?Curr Health Sci J. 2020 Jan-Mar;46(1):5-10. doi: 10.12865/CHSJ.46.01.01. Epub 2020 Mar 31. Curr Health Sci J. 2020. PMID: 32637159 Free PMC article. Review.
Cited by
-
Gene Expression Profile Analysis of Human Epidermal Keratinocytes Expressing Human Papillomavirus Type 8 E7.Pathol Oncol Res. 2022 May 18;28:1610176. doi: 10.3389/pore.2022.1610176. eCollection 2022. Pathol Oncol Res. 2022. PMID: 35665406 Free PMC article.
-
Identification of Differentially Methylated CpG Sites in Fibroblasts from Keloid Scars.Biomedicines. 2020 Jun 28;8(7):181. doi: 10.3390/biomedicines8070181. Biomedicines. 2020. PMID: 32605309 Free PMC article.
-
Global gene methylation profiling of common warts caused by human papillomaviruses infection.Saudi J Biol Sci. 2021 Jan;28(1):612-622. doi: 10.1016/j.sjbs.2020.10.050. Epub 2020 Nov 2. Saudi J Biol Sci. 2021. PMID: 33424347 Free PMC article.
References
-
- Lim DHK, Maher ER. DNA methylation: a form of epigenetic control of gene expression. Obstet Gynaecol. 2010;12:37–42.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

