Cyclic AMP-CRP Modulates the Cell Morphology of Klebsiella pneumoniae in High-Glucose Environment
- PMID: 32038513
- PMCID: PMC6985210
- DOI: 10.3389/fmicb.2019.02984
Cyclic AMP-CRP Modulates the Cell Morphology of Klebsiella pneumoniae in High-Glucose Environment
Abstract
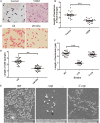
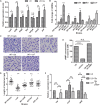
Bacteria can modify their morphology in response to environmental stimuli for survival or host defense evasion. The rich glucose in vivo or in the Luria-Bertani (LB) medium shortened the cell length of Klebsiella pneumoniae. The environmental glucose decreased the levels of cyclic AMP (cAMP) and the transcription of crp, which declined the cAMP-cAMP receptor protein (cAMP-CRP) activity. The cell length of crp deletion mutant was significantly shorter than that of the wild type (0.981 ± 0.057 μm vs. 2.415 ± 0.075 μm, P < 0.001). These results indicated that the high environmental glucose alters the bacterial morphology to a round form through regulating the activity of cAMP-CRP complex. Comparative proteomics analysis showed increased expression of 10 proteins involved in cell division or cell wall biosynthesis in the crp deletion strain. Five of them (ompA, tolB, ybgC, ftsI, and rcsF) were selected to verify their expression in the high-glucose environment, and overexpression of tolB or rcsF shortened the bacterial length similar to that of the crp deletion strain. Electrophoretic mobility shift assay indicated that CRP directly negatively regulates the transcription of tolB and rcsF by binding to the promoter regions. This study first proved the role and partial regulation mechanism of CRP in altering cell morphology during infection and provided a theoretical basis for elucidating the mechanism in diabetes mellitus susceptible to K. pneumoniae.
Keywords: Klebsiella pneumoniae; cell morphology; cyclic AMP-CRP; glucose; regulation.
Copyright © 2020 Liu, Li, Xu, Wang, Li, Yuan, Wang, Yang and Li.
Figures


Similar articles
-
Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae.PLoS One. 2013;8(2):e54430. doi: 10.1371/journal.pone.0054430. Epub 2013 Feb 11. PLoS One. 2013. PMID: 23408939 Free PMC article.
-
CRP-Cyclic AMP Regulates the Expression of Type 3 Fimbriae via Cyclic di-GMP in Klebsiella pneumoniae.PLoS One. 2016 Sep 15;11(9):e0162884. doi: 10.1371/journal.pone.0162884. eCollection 2016. PLoS One. 2016. PMID: 27631471 Free PMC article.
-
EtcABC, a Putative EII Complex, Regulates Type 3 Fimbriae via CRP-cAMP Signaling in Klebsiella pneumoniae.Front Microbiol. 2019 Jul 9;10:1558. doi: 10.3389/fmicb.2019.01558. eCollection 2019. Front Microbiol. 2019. PMID: 31354661 Free PMC article.
-
Syn, anti, and finally both conformations of cyclic AMP are involved in the CRP-dependent transcription initiation mechanism in E. coli lac operon.Cell Biochem Funct. 2008 Jun;26(4):399-405. doi: 10.1002/cbf.1462. Cell Biochem Funct. 2008. PMID: 18338329 Review.
-
Rethinking the roles of CRP, cAMP, and sugar-mediated global regulation in the Vibrionaceae.Curr Genet. 2016 Feb;62(1):39-45. doi: 10.1007/s00294-015-0508-8. Epub 2015 Jul 28. Curr Genet. 2016. PMID: 26215147 Review.
Cited by
-
Blindness in Right Eyes after Enema: A Case of Klebsiella pneumoniae-Related Invasive Liver Abscess Syndrome with Endophthalmitis-Caused Blindness as the First Symptom.Case Rep Med. 2024 Feb 13;2024:5573160. doi: 10.1155/2024/5573160. eCollection 2024. Case Rep Med. 2024. PMID: 38380356 Free PMC article.
-
[Construction and characterization of a modA gene mutant strain of Klebsiella pneumoniae].Nan Fang Yi Ke Da Xue Xue Bao. 2024 Apr 20;44(4):748-756. doi: 10.12122/j.issn.1673-4254.2024.04.17. Nan Fang Yi Ke Da Xue Xue Bao. 2024. PMID: 38708509 Free PMC article. Chinese.
-
Retrospective study of characteristics and management of pyogenic liver abscess during 5 years' experience.Int J Clin Exp Pathol. 2021 Feb 1;14(2):252-260. eCollection 2021. Int J Clin Exp Pathol. 2021. PMID: 33564358 Free PMC article.
-
Relevance of the Adjuvant Effect between Cellular Homeostasis and Resistance to Antibiotics in Gram-Negative Bacteria with Pathogenic Capacity: A Study of Klebsiella pneumoniae.Antibiotics (Basel). 2024 May 26;13(6):490. doi: 10.3390/antibiotics13060490. Antibiotics (Basel). 2024. PMID: 38927157 Free PMC article. Review.
-
Intestinal carbapenem-resistant Klebsiella pneumoniae undergoes complex transcriptional reprogramming following immune activation.Gut Microbes. 2024 Jan-Dec;16(1):2340486. doi: 10.1080/19490976.2024.2340486. Epub 2024 Apr 24. Gut Microbes. 2024. PMID: 38659243 Free PMC article.
References
-
- Bryan S. J., Burroughs N. J., Evered C., Sacharz J., Nenninger A., Mullineaux C. W., et al. (2011). Loss of the SPHF homologue Slr1768 leads to a catastrophic failure in the maintenance of thylakoid membranes in Synechocystis sp. PCC 6803. PLoS One 6:e19625. 10.1371/journal.pone.0019625 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

