Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion
- PMID: 32038698
- PMCID: PMC6992662
- DOI: 10.3389/fpls.2019.01741
Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion
Abstract
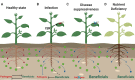
Plants host a mesmerizing diversity of microbes inside and around their roots, known as the microbiome. The microbiome is composed mostly of fungi, bacteria, oomycetes, and archaea that can be either pathogenic or beneficial for plant health and fitness. To grow healthy, plants need to surveil soil niches around the roots for the detection of pathogenic microbes, and in parallel maximize the services of beneficial microbes in nutrients uptake and growth promotion. Plants employ a palette of mechanisms to modulate their microbiome including structural modifications, the exudation of secondary metabolites and the coordinated action of different defence responses. Here, we review the current understanding on the composition and activity of the root microbiome and how different plant molecules can shape the structure of the root-associated microbial communities. Examples are given on interactions that occur in the rhizosphere between plants and soilborne fungi. We also present some well-established examples of microbiome harnessing to highlight how plants can maximize their fitness by selecting their microbiome. Understanding how plants manipulate their microbiome can aid in the design of next-generation microbial inoculants for targeted disease suppression and enhanced plant growth.
Keywords: disease suppression; microbial inoculants; microbiota; plant defense; plant growth promotion; plant molecules; root exudation; root microbiome.
Copyright © 2020 Pascale, Proietti, Pantelides and Stringlis.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

