StABI5 Involved in the Regulation of Chloroplast Development and Photosynthesis in Potato
- PMID: 32041112
- PMCID: PMC7036812
- DOI: 10.3390/ijms21031068
StABI5 Involved in the Regulation of Chloroplast Development and Photosynthesis in Potato
Abstract
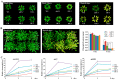
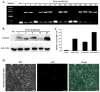
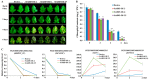
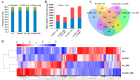
Abscisic acid (ABA) insensitive 5 (ABI5)-a core transcription factor of the ABA signaling pathway-is a basic leucine zipper transcription factor that plays a key role in the regulation of seed germination and early seedling growth. ABI5 interacts with other phytohormone signals to regulate plant growth and development, and stress responses in Arabidopsis, but little is known about the functions of ABI5 in potatoes. Here, we find that StABI5 is involved in the regulation of chloroplast development and photosynthesis. Genetic analysis indicates that StABI5 overexpression transgenic potato lines accelerate dark-induced leaf yellowing and senescence. The chlorophyll contents of overexpressed StABI5 transgenic potato lines were significantly decreased in comparison to those of wild-type Desiree potatoes under dark conditions. Additionally, the RNA-sequencing (RNA-seq) analysis shows that many metabolic processes are changed in overexpressed StABI5 transgenic potatoes. Most of the genes involved in photosynthesis and carbon fixation are significantly down-regulated, especially the chlorophyll a-b binding protein, photosystem I, and photosystem II. These observations indicate that StABI5 negatively regulates chloroplast development and photosynthesis, and provides some insights into the functions of StABI5 in regard to potato growth.
Keywords: StABI5; chlorophyll; chloroplast development; photosynthesis; potato.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Ectopic expression of a hot pepper bZIP-like transcription factor in potato enhances drought tolerance without decreasing tuber yield.Plant Mol Biol. 2015 Nov;89(4-5):421-31. doi: 10.1007/s11103-015-0378-y. Epub 2015 Sep 22. Plant Mol Biol. 2015. PMID: 26394867
-
Potato Annexin STANN1 Promotes Drought Tolerance and Mitigates Light Stress in Transgenic Solanum tuberosum L. Plants.PLoS One. 2015 Jul 14;10(7):e0132683. doi: 10.1371/journal.pone.0132683. eCollection 2015. PLoS One. 2015. PMID: 26172952 Free PMC article.
-
Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development.Plant J. 2014 Nov;80(4):654-68. doi: 10.1111/tpj.12670. Plant J. 2014. PMID: 25231920
-
BBX21 reduces abscisic acid sensitivity, mesophyll conductance and chloroplast electron transport capacity to increase photosynthesis and water use efficiency in potato plants cultivated under moderated drought.Plant J. 2021 Nov;108(4):1131-1144. doi: 10.1111/tpj.15499. Epub 2021 Oct 18. Plant J. 2021. PMID: 34606658
-
Regulation of Chloroplast Development and Function at Adverse Temperatures in Plants.Plant Cell Physiol. 2022 May 16;63(5):580-591. doi: 10.1093/pcp/pcac022. Plant Cell Physiol. 2022. PMID: 35141744 Review.
Cited by
-
ABI5 promotes heat stress-induced chlorophyll degradation by modulating the stability of MYB44 in cucumber.Hortic Res. 2023 May 4;10(6):uhad089. doi: 10.1093/hr/uhad089. eCollection 2023 Jun. Hortic Res. 2023. PMID: 37334179 Free PMC article.
-
Prunus persica Terpene Synthase PpTPS1 Interacts with PpABI5 to Enhance Salt Resistance in Transgenic Tomatoes.Front Plant Sci. 2022 Feb 23;13:807342. doi: 10.3389/fpls.2022.807342. eCollection 2022. Front Plant Sci. 2022. PMID: 35283925 Free PMC article.
-
Volatile Compounds From Bacillus, Serratia, and Pseudomonas Promote Growth and Alter the Transcriptional Landscape of Solanum tuberosum in a Passively Ventilated Growth System.Front Microbiol. 2021 Jul 21;12:628437. doi: 10.3389/fmicb.2021.628437. eCollection 2021. Front Microbiol. 2021. PMID: 34367077 Free PMC article.
-
The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2.Hortic Res. 2021 Jul 1;8(1):159. doi: 10.1038/s41438-021-00593-0. Hortic Res. 2021. PMID: 34193837 Free PMC article.
-
Genome-Wide Identification of 14-3-3 gene family reveals their diverse responses to abiotic stress by interacting with StABI5 in Potato (Solanum tuberosum L.).Front Plant Sci. 2023 Jan 9;13:1090571. doi: 10.3389/fpls.2022.1090571. eCollection 2022. Front Plant Sci. 2023. PMID: 36699847 Free PMC article.
References
-
- Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R.K., Kumar V., Verma R., Upadhyay R.G., Pandey M., et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017;8:161. doi: 10.3389/fpls.2017.00161. - DOI - PMC - PubMed
-
- Dong T., Park Y., Hwang I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015;58:29–48. - PubMed
-
- Finkelstein R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. doi: 10.1046/j.1365-313X.1994.5060765.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

