Analysis of the Bacterial Diversity of Paipa Cheese (a Traditional Raw Cow's Milk Cheese from Colombia) by High-Throughput Sequencing
- PMID: 32041151
- PMCID: PMC7074763
- DOI: 10.3390/microorganisms8020218
Analysis of the Bacterial Diversity of Paipa Cheese (a Traditional Raw Cow's Milk Cheese from Colombia) by High-Throughput Sequencing
Abstract
Background: Paipa cheese is a traditional, semi-ripened cheese made from raw cow's milk in Colombia. The aim of this work was to gain insights on the microbiota of Paipa cheese by using a culture-independent approach.
Method: two batches of Paipa cheese from three formal producers were sampled during ripening for 28 days. Total DNA from the cheese samples was used to obtain 16S rRNA gene sequences by using Illumina technology.
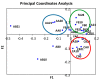
Results: Firmicutes was the main phylum found in the cheeses (relative abundances: 59.2-82.0%), followed by Proteobacteria, Actinobacteria and Bacteroidetes. Lactococcus was the main genus, but other lactic acid bacteria (Enterococcus, Leuconostoc and Streptococcus) were also detected. Stapylococcus was also relevant in some cheese samples. The most important Proteobacteria were Enterobacteriaceae, Aeromonadaceae and Moraxellaceae. Enterobacter and Enterobacteriaceae (others) were detected in all cheese samples. Serratia and Citrobacter were detected in some samples. Aeromonas and Acinetobacter were also relevant. Other minor genera detected were Marinomonas, Corynebacterium 1 and Chryseobacterium. The principal coordinates analysis suggested that there were producer-dependent differences in the microbiota of Paipa cheeses.
Conclusions: lactic acid bacteria are the main bacterial group in Paipa cheeses. However, other bacterial groups, including spoilage bacteria, potentially toxin producers, and bacteria potentially pathogenic to humans and/or prone to carry antimicrobial resistance genes are also relevant in the cheeses.
Keywords: Aromonadaceae; Enterobacteriaceae; Moraxellaceae; Paipa cheese; Staphylococcus; bacterial diversity; lactic acid bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Microbial shifts through the ripening of the "Entre Serras" Minas artisanal cheese monitored by high-throughput sequencing.Food Res Int. 2021 Jan;139:109803. doi: 10.1016/j.foodres.2020.109803. Epub 2020 Oct 24. Food Res Int. 2021. PMID: 33509447
-
The microbiota of high-moisture mozzarella cheese produced with different acidification methods.Int J Food Microbiol. 2016 Jan 4;216:9-17. doi: 10.1016/j.ijfoodmicro.2015.09.002. Epub 2015 Sep 9. Int J Food Microbiol. 2016. PMID: 26384211
-
In-Depth Investigation of the Safety of Wooden Shelves Used for Traditional Cheese Ripening.Appl Environ Microbiol. 2021 Nov 10;87(23):e0152421. doi: 10.1128/AEM.01524-21. Epub 2021 Sep 22. Appl Environ Microbiol. 2021. PMID: 34550766 Free PMC article.
-
Traditional cheeses: rich and diverse microbiota with associated benefits.Int J Food Microbiol. 2014 May 2;177:136-54. doi: 10.1016/j.ijfoodmicro.2014.02.019. Epub 2014 Mar 3. Int J Food Microbiol. 2014. PMID: 24642348 Review.
-
The Microfloras and Sensory Profiles of Selected Protected Designation of Origin Italian Cheeses.Microbiol Spectr. 2014 Feb;2(1):CM-0007-2012. doi: 10.1128/microbiolspec.CM-0007-2012. Microbiol Spectr. 2014. PMID: 26082116 Review.
Cited by
-
Microbial Interactions within the Cheese Ecosystem and Their Application to Improve Quality and Safety.Foods. 2021 Mar 12;10(3):602. doi: 10.3390/foods10030602. Foods. 2021. PMID: 33809159 Free PMC article. Review.
-
Feasibility of Using Carvacrol/Starch Edible Coatings to Improve the Quality of Paipa Cheese.Polymers (Basel). 2021 Jul 30;13(15):2516. doi: 10.3390/polym13152516. Polymers (Basel). 2021. PMID: 34372119 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

