Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B
- PMID: 32041218
- PMCID: PMC7158668
- DOI: 10.3390/vaccines8010070
Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B
Abstract
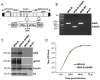
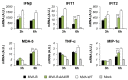
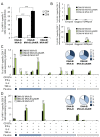
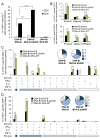
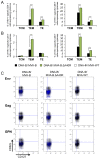
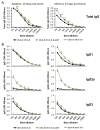
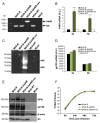
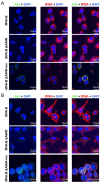
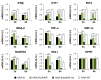
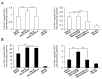
Development of a safe and efficacious vaccine against the HIV/AIDS pandemic remains a major scientific goal. We previously described an HIV/AIDS vaccine based on the modified vaccinia virus Ankara (MVA) expressing HIV-1 gp120 and Gag-Pol-Nef (GPN) of clade B (termed MVA-B), which showed moderate immunogenicity in phase I prophylactic and therapeutic clinical trials. Here, to improve the immunogenicity of MVA-B, we generated a novel recombinant virus, MVA-B ΔA40R, by deleting in the MVA-B genome the vaccinia virus (VACV) A40R gene, which encodes a protein with unknown immune function. The innate immune responses triggered by MVA-B ΔA40R in infected human macrophages, in comparison to parental MVA-B, revealed an increase in the mRNA expression levels of interferon (IFN)-β, IFN-induced genes, and chemokines. Compared to priming with DNA-B (a mixture of DNA-gp120 plus DNA-GPN) and boosting with MVA-B, mice immunized with a DNA-B/MVA-B ΔA40R regimen induced higher magnitude of adaptive and memory HIV-1-specific CD4+ and CD8+ T-cell immune responses that were highly polyfunctional, mainly directed against Env. and of an effector memory phenotype, together with enhanced levels of antibodies against HIV-1 gp120. Reintroduction of the A40R gene into the MVA-B ΔA40R genome (virus termed MVA-B ΔA40R-rev) promoted in infected cells high mRNA and protein A40 levels, with A40 protein localized in the cell membrane. MVA-B ΔA40R-rev significantly reduced mRNA levels of IFN-β and of several other innate immune-related genes in infected human macrophages. In immunized mice, MVA-B ΔA40R-rev reduced the magnitude of the HIV-1-specific CD4+ and CD8+ T cell responses compared to MVA-B ΔA40R. These results revealed an immunosuppressive role of the A40 protein, findings relevant for the optimization of poxvirus vectors as vaccines.
Keywords: A40R gene; HIV vaccine; MVA; immune responses; mice; poxvirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Barouch D.H., Santra S., Schmitz J.E., Kuroda M.J., Fu T.M., Wagner W., Bilska M., Craiu A., Zheng X.X., Krivulka G.R., et al. Control of viremia prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. - DOI - PubMed
-
- Mooij P., Balla-Jhagjhoorsingh S.S., Koopman G., Beenhakker N., van Haaften P., Baak I., Nieuwenhuis I.G., Kondova I., Wagner R., Wolf H., et al. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus Type 1 vaccine candidates provide comparable efficacies in Primates. J. Virol. 2008;82:2975–2988. doi: 10.1128/JVI.02216-07. - DOI - PMC - PubMed
-
- Amara R.R., Ibegbu C., Villinger F., Montefiori D.C., Sharma S., Nigam P., Xu Y., McClure H.M., Robinson H.L. Studies using a viral challenge and CD8 T cell depletions on the roles of cellular and humoral immunity in the control of an SHIV-89.6P challenge in DNA/MVA-vaccinated macaques. Virology. 2005;343:246–255. doi: 10.1016/j.virol.2005.08.027. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

