Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay
- PMID: 32041247
- PMCID: PMC7037193
- DOI: 10.3390/ijms21031085
Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay
Abstract
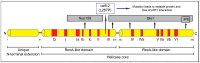
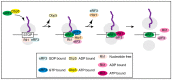
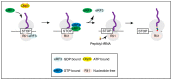
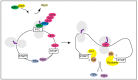
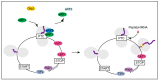
The DEAD-box protein Dbp5 (human DDX19) remodels RNA-protein complexes. Dbp5 functions in ribonucleoprotein export and translation termination. Termination occurs, when the ribosome has reached a stop codon through the Dbp5 mediated delivery of the eukaryotic termination factor eRF1. eRF1 contacts eRF3 upon dissociation of Dbp5, resulting in polypeptide chain release and subsequent ribosomal subunit splitting. Mutations in DBP5 lead to stop codon readthrough, because the eRF1 and eRF3 interaction is not controlled and occurs prematurely. This identifies Dbp5/DDX19 as a possible potent drug target for nonsense suppression therapy. Neurodegenerative diseases and cancer are caused in many cases by the loss of a gene product, because its mRNA contained a premature termination codon (PTC) and is thus eliminated through the nonsense mediated decay (NMD) pathway, which is described in the second half of this review. We discuss translation termination and NMD in the light of Dbp5/DDX19 and subsequently speculate on reducing Dbp5/DDX19 activity to allow readthrough of the PTC and production of a full-length protein to detract the RNA from NMD as a possible treatment for diseases.
Keywords: DDX19; Dbp5; NMD; Rat8; mRNA degradation; mRNA quality control; translation; translation termination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3.Nucleic Acids Res. 2019 May 21;47(9):4798-4813. doi: 10.1093/nar/gkz177. Nucleic Acids Res. 2019. PMID: 30873535 Free PMC article.
-
The DEAD-box RNA helicase Dbp5 functions in translation termination.Science. 2007 Feb 2;315(5812):646-9. doi: 10.1126/science.1134641. Science. 2007. PMID: 17272721
-
RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes.Nucleic Acids Res. 2017 Feb 17;45(3):1307-1318. doi: 10.1093/nar/gkw1239. Nucleic Acids Res. 2017. PMID: 28180304 Free PMC article.
-
The Meaning of NMD: Translate or Perish.Trends Genet. 2016 Jul;32(7):395-407. doi: 10.1016/j.tig.2016.04.007. Epub 2016 May 14. Trends Genet. 2016. PMID: 27185236 Review.
-
Dbp5 - from nuclear export to translation.Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
-
FOXC2 Disease Mutations Identified in Lymphedema Distichiasis Patients Impair Transcriptional Activity and Cell Proliferation.Int J Mol Sci. 2020 Jul 20;21(14):5112. doi: 10.3390/ijms21145112. Int J Mol Sci. 2020. PMID: 32698337 Free PMC article.
-
Gle1 is required for tRNA to stimulate Dbp5 ATPase activity in vitro and to promote Dbp5 mediated tRNA export in vivo.bioRxiv [Preprint]. 2023 Nov 9:2023.06.29.547072. doi: 10.1101/2023.06.29.547072. bioRxiv. 2023. Update in: Elife. 2024 Jan 08;12:RP89835. doi: 10.7554/eLife.89835. PMID: 37425677 Free PMC article. Updated. Preprint.
-
Protein translation: biological processes and therapeutic strategies for human diseases.Signal Transduct Target Ther. 2024 Feb 23;9(1):44. doi: 10.1038/s41392-024-01749-9. Signal Transduct Target Ther. 2024. PMID: 38388452 Free PMC article. Review.
-
Emerging molecular functions and novel roles for the DEAD-box protein Dbp5/DDX19 in gene expression.Cell Mol Life Sci. 2021 Mar;78(5):2019-2030. doi: 10.1007/s00018-020-03680-y. Epub 2020 Nov 17. Cell Mol Life Sci. 2021. PMID: 33205304 Free PMC article. Review.
-
Functional Activity of Isoform 2 of Human eRF1.Int J Mol Sci. 2024 Jul 22;25(14):7997. doi: 10.3390/ijms25147997. Int J Mol Sci. 2024. PMID: 39063238 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

