A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases
- PMID: 32041280
- PMCID: PMC7037290
- DOI: 10.3390/ijerph17031035
A Network-Based Bioinformatics Approach to Identify Molecular Biomarkers for Type 2 Diabetes that Are Linked to the Progression of Neurological Diseases
Abstract
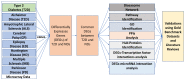
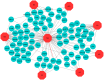
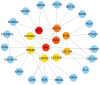
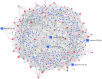
Neurological diseases (NDs) are progressive disorders, the progression of which can be significantly affected by a range of common diseases that present as comorbidities. Clinical studies, including epidemiological and neuropathological analyses, indicate that patients with type 2 diabetes (T2D) have worse progression of NDs, suggesting pathogenic links between NDs and T2D. However, finding causal or predisposing factors that link T2D and NDs remains challenging. To address these problems, we developed a high-throughput network-based quantitative pipeline using agnostic approaches to identify genes expressed abnormally in both T2D and NDs, to identify some of the shared molecular pathways that may underpin T2D and ND interaction. We employed gene expression transcriptomic datasets from control and disease-affected individuals and identified differentially expressed genes (DEGs) in tissues of patients with T2D and ND when compared to unaffected control individuals. One hundred and ninety seven DEGs (99 up-regulated and 98 down-regulated in affected individuals) that were common to both the T2D and the ND datasets were identified. Functional annotation of these identified DEGs revealed the involvement of significant cell signaling associated molecular pathways. The overlapping DEGs (i.e., seen in both T2D and ND datasets) were then used to extract the most significant GO terms. We performed validation of these results with gold benchmark databases and literature searching, which identified which genes and pathways had been previously linked to NDs or T2D and which are novel. Hub proteins in the pathways were identified (including DNM2, DNM1, MYH14, PACSIN2, TFRC, PDE4D, ENTPD1, PLK4, CDC20B, and CDC14A) using protein-protein interaction analysis which have not previously been described as playing a role in these diseases. To reveal the transcriptional and post-transcriptional regulators of the DEGs we used transcription factor (TF) interactions analysis and DEG-microRNAs (miRNAs) interaction analysis, respectively. We thus identified the following TFs as important in driving expression of our T2D/ND common genes: FOXC1, GATA2, FOXL1, YY1, E2F1, NFIC, NFYA, USF2, HINFP, MEF2A, SRF, NFKB1, USF2, HINFP, MEF2A, SRF, NFKB1, PDE4D, CREB1, SP1, HOXA5, SREBF1, TFAP2A, STAT3, POU2F2, TP53, PPARG, and JUN. MicroRNAs that affect expression of these genes include mir-335-5p, mir-16-5p, mir-93-5p, mir-17-5p, mir-124-3p. Thus, our transcriptomic data analysis identifies novel potential links between NDs and T2D pathologies that may underlie comorbidity interactions, links that may include potential targets for therapeutic intervention. In sum, our neighborhood-based benchmarking and multilayer network topology methods identified novel putative biomarkers that indicate how type 2 diabetes (T2D) and these neurological diseases interact and pathways that, in the future, may be targeted for treatment.
Keywords: bioinformatics; computational biology; gene ontology; neurological disease; pathways; protein; type 2 diabetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Network-Based Genetic Profiling Reveals Cellular Pathway Differences Between Follicular Thyroid Carcinoma and Follicular Thyroid Adenoma.Int J Environ Res Public Health. 2020 Feb 20;17(4):1373. doi: 10.3390/ijerph17041373. Int J Environ Res Public Health. 2020. PMID: 32093341 Free PMC article.
-
Bioinformatics and Systems Biology Approaches to Identify the Synergistic Effects of Alcohol Use Disorder on the Progression of Neurological Diseases.Neuroscience. 2024 Apr 5;543:65-82. doi: 10.1016/j.neuroscience.2024.02.015. Epub 2024 Feb 23. Neuroscience. 2024. PMID: 38401711
-
Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: Insights from a systems biomedicine perspective.Genomics. 2020 Mar;112(2):1290-1299. doi: 10.1016/j.ygeno.2019.07.018. Epub 2019 Aug 1. Genomics. 2020. PMID: 31377428
-
Exploration of key drug target proteins highlighting their related regulatory molecules, functional pathways and drug candidates associated with delirium: evidence from meta-data analyses.BMC Geriatr. 2023 Nov 22;23(1):767. doi: 10.1186/s12877-023-04457-1. BMC Geriatr. 2023. PMID: 37993790 Free PMC article.
-
Systematic bioinformatic analysis of nutrigenomic data of flavanols in cell models of cardiometabolic disease.Food Funct. 2020 Jun 24;11(6):5040-5064. doi: 10.1039/d0fo00701c. Food Funct. 2020. PMID: 32537624
Cited by
-
circDYRK1A tethers biological behaviors of gastric carcinoma using novel bioinformatics analysis and experimental validations.Sci Rep. 2023 May 22;13(1):8265. doi: 10.1038/s41598-023-33861-1. Sci Rep. 2023. PMID: 37217530 Free PMC article.
-
An integrated complete-genome sequencing and systems biology approach to predict antimicrobial resistance genes in the virulent bacterial strains of Moraxella catarrhalis.Brief Funct Genomics. 2023 Jul 17;22(4):375-391. doi: 10.1093/bfgp/elad005. Brief Funct Genomics. 2023. PMID: 36881677 Free PMC article.
-
qPCR Analysis Reveals Association of Differential Expression of SRR, NFKB1, and PDE4B Genes With Type 2 Diabetes Mellitus.Front Endocrinol (Lausanne). 2022 Jan 3;12:774696. doi: 10.3389/fendo.2021.774696. eCollection 2021. Front Endocrinol (Lausanne). 2022. PMID: 35046895 Free PMC article.
-
Systems biology and in silico-based analysis of PCOS revealed the risk of metabolic disorders.Heliyon. 2022 Dec 22;8(12):e12480. doi: 10.1016/j.heliyon.2022.e12480. eCollection 2022 Dec. Heliyon. 2022. PMID: 36619413 Free PMC article.
-
Bioinformatics and In silico approaches to identify novel biomarkers and key pathways for cancers that are linked to the progression of female infertility: A comprehensive approach for drug discovery.PLoS One. 2023 Jan 6;18(1):e0265746. doi: 10.1371/journal.pone.0265746. eCollection 2023. PLoS One. 2023. PMID: 36608061 Free PMC article.
References
-
- Mallorquí-Bagué N., Lozano-Madrid M., Toledo E., Corella D., Salas-Salvadó J., Cuenca-Royo A., Vioque J., Romaguera D., Martínez J.A., Wärnberg J., et al. Type 2 diabetes and cognitive impairment in an older population with overweight or obesity and metabolic syndrome: Baseline cross-sectional analysis of the predimed-plus study. Sci. Rep. 2018;8:16128. doi: 10.1038/s41598-018-33843-8. - DOI - PMC - PubMed
-
- American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27:S5eS10. - PubMed
-
- Xu L., Zhao H., Qiu J., Zhu W., Lei H., Cai Z., Lin W.H., Huang W., Zhang H., Zhang Y.T. The different effects of BMI and WC on organ damage in patients from a cardiac rehabilitation program after acute coronary syndrome. BioMed Res. Int. 2015;2015:942695. doi: 10.1155/2015/942695. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

