The Three-Dimensional Ultrastructure of the Human Alveolar Epithelium Revealed by Focused Ion Beam Electron Microscopy
- PMID: 32041332
- PMCID: PMC7038159
- DOI: 10.3390/ijms21031089
The Three-Dimensional Ultrastructure of the Human Alveolar Epithelium Revealed by Focused Ion Beam Electron Microscopy
Abstract
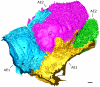
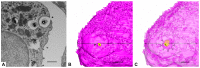
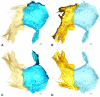
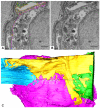
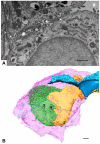
Thin type 1 alveolar epithelial (AE1) and surfactant producing type 2 alveolar epithelial (AE2) cells line the alveoli in the lung and are essential for normal lung function. Function is intimately interrelated to structure, so that detailed knowledge of the epithelial ultrastructure can significantly enhance our understanding of its function. The basolateral surface of the cells or the epithelial contact sites are of special interest, because they play an important role in intercellular communication or stabilizing the epithelium. The latter is in particular important for the lung with its variable volume. The aim of the present study was to investigate the three-dimensional (3D) ultrastructure of the human alveolar epithelium focusing on contact sites and the basolateral cell membrane of AE2 cells using focused ion beam electron microscopy and subsequent 3D reconstructions. The study provides detailed surface reconstructions of two AE1 cell domains and two AE2 cells, showing AE1/AE1, AE1/AE2 and AE2/AE2 contact sites, basolateral microvilli pits at AE2 cells and small AE1 processes beneath AE2 cells. Furthermore, we show reconstructions of a surfactant secretion pore, enlargements of the apical AE1 cell surface and long folds bordering grooves on the basal AE1 cell surface. The functional implications of our findings are discussed. These findings may lay the structural basis for further molecular investigations.
Keywords: 3D reconstruction; alveolus; focused ion beam scanning electron microscopy; lung; type 1 alveolar epithelial cell; type 2 alveolar epithelial cell.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, nor in the decision to publish the results.
Figures







Similar articles
-
Alveolar epithelial type II cell: defender of the alveolus revisited.Respir Res. 2001;2(1):33-46. doi: 10.1186/rr36. Epub 2001 Jan 15. Respir Res. 2001. PMID: 11686863 Free PMC article. Review.
-
Air-blood barrier thickening and alterations of alveolar epithelial type 2 cells in mouse lungs with disrupted hepcidin/ferroportin regulatory system.Histochem Cell Biol. 2019 Mar;151(3):217-228. doi: 10.1007/s00418-018-1737-y. Epub 2018 Oct 3. Histochem Cell Biol. 2019. PMID: 30280242
-
Cl(-)-HCO3- exchanger isoform AE2 is restricted to the basolateral surface of alveolar epithelial cell monolayers.Am J Respir Cell Mol Biol. 1995 Feb;12(2):211-9. doi: 10.1165/ajrcmb.12.2.7865219. Am J Respir Cell Mol Biol. 1995. PMID: 7865219
-
Secretion of surfactant by rat alveolar type II cells: morphometric analysis and three-dimensional reconstruction.Exp Lung Res. 1985;9(3-4):351-61. doi: 10.3109/01902148509057533. Exp Lung Res. 1985. PMID: 3841051
-
Chloride and potassium channel function in alveolar epithelial cells.Am J Physiol Lung Cell Mol Physiol. 2003 May;284(5):L689-700. doi: 10.1152/ajplung.00256.2002. Am J Physiol Lung Cell Mol Physiol. 2003. PMID: 12676759 Review.
Cited by
-
Ultrastructural 3D Microscopy for Biomedicine: Principles, Applications, and Perspectives.Acta Naturae. 2024 Jan-Mar;16(1):14-29. doi: 10.32607/actanaturae.27323. Acta Naturae. 2024. PMID: 38698961 Free PMC article.
-
How innovations in methodology offer new prospects for volume electron microscopy.J Microsc. 2022 Sep;287(3):114-137. doi: 10.1111/jmi.13134. Epub 2022 Jul 27. J Microsc. 2022. PMID: 35810393 Free PMC article. Review.
-
Biological volume EM with focused Ga ion beam depends on formation of radiation-resistant Ga-rich layer at block face.bioRxiv [Preprint]. 2025 Mar 12:2024.09.16.613321. doi: 10.1101/2024.09.16.613321. bioRxiv. 2025. PMID: 39803553 Free PMC article. Preprint.
-
Volume electron microscopy: analyzing the lung.Histochem Cell Biol. 2021 Feb;155(2):241-260. doi: 10.1007/s00418-020-01916-3. Epub 2020 Sep 17. Histochem Cell Biol. 2021. PMID: 32944795 Free PMC article. Review.
-
Distribution and volume of mitochondria in alveolar epithelial type 1 cells in infant and adult human lungs.Histochem Cell Biol. 2024 Nov 18;163(1):7. doi: 10.1007/s00418-024-02332-7. Histochem Cell Biol. 2024. PMID: 39557665 Free PMC article.
References
-
- Ochs M., Weibel E.R. Functional design of the human lung for gas exchange. In: Fishman A.P., Elias J.A., Fishman J.A., Grippi M.A., Senior R.M., Pack A.I., editors. Fishman’s Pulmonary Diseases and Disorders. 4th ed. Volume 1. McGraw-Hill Medical; New York, NY, USA: 2008. pp. 23–69. Chapter 2.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

